6CSU
 
 | | The structure of the Cep63-Cep152 heterotetrameric complex | | Descriptor: | Centrosomal protein of 152 kDa, Centrosomal protein of 63 kDa | | Authors: | Lee, E, Chen, Y, Zhang, L, Kim, T.S, Ahn, J.I, Park, J.E, Lee, K.S. | | Deposit date: | 2018-03-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular architecture of a cylindrical self-assembly at human centrosomes.
Nat Commun, 10, 2019
|
|
6LFK
 
 | |
7C4P
 
 | | Crystal structure of DBD plasma treated zebrafish TRF2 myb-domain complexed with DNA | | Descriptor: | DNA (5'-D(*CP*CP*CP*TP*AP*AP*CP*CP*CP*TP*AP*A)-3'), DNA (5'-D(*TP*TP*AP*GP*GP*GP*TP*TP*AP*G)-3'), DNA (5'-D(*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*G)-3'), ... | | Authors: | Jin, Z, Park, J.H, Yun, J.H, Park, S.Y, Lee, W. | | Deposit date: | 2020-05-18 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Crystal structure of DBD plasma treated zebrafish TRF2 myb-domain complexed with DNA
To Be Published
|
|
7C4Q
 
 | | Crystal structure of DBD plasma treated zebrafish TRF2 myb-domain complexed with DNA | | Descriptor: | DNA (5'-D(*CP*CP*CP*TP*AP*AP*CP*CP*CP*TP*AP*A)-3'), DNA (5'-D(*TP*TP*AP*GP*GP*GP*TP*TP*AP*G)-3'), DNA (5'-D(*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*G)-3'), ... | | Authors: | Jin, Z, Park, J.H, Yun, J.H, Park, S.Y, Lee, W. | | Deposit date: | 2020-05-18 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of DBD plasma treated zebrafish TRF2 myb-domain complexed with DNA
To Be Published
|
|
6KZ7
 
 | | The crystal structure of BAF155 SWIRM domain and N-terminal elongated hSNF5 RPT1 domain complex: Chromatin remodeling complex | | Descriptor: | SWI/SNF complex subunit SMARCC1, SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1 | | Authors: | Lee, W, Han, J, Kim, I, Park, J.H, Joo, K, Lee, J, Suh, J.Y. | | Deposit date: | 2019-09-23 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | A Coil-to-Helix Transition Serves as a Binding Motif for hSNF5 and BAF155 Interaction.
Int J Mol Sci, 21, 2020
|
|
6J43
 
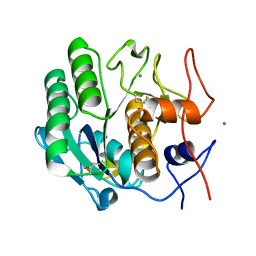 | | Proteinase K determined by PAL-XFEL | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Lee, S.J, Park, J. | | Deposit date: | 2019-01-07 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Application of a high-throughput microcrystal delivery system to serial femtosecond crystallography.
J.Appl.Crystallogr., 53, 2020
|
|
2R64
 
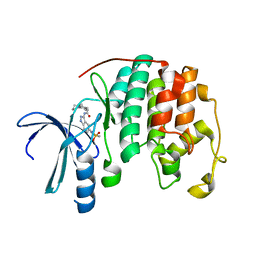 | | Crystal structure of a 3-aminoindazole compound with CDK2 | | Descriptor: | Cell division protein kinase 2, N-[5-(1,1-DIOXIDOISOTHIAZOLIDIN-2-YL)-1H-INDAZOL-3-YL]-2-(4-PIPERIDIN-1-YLPHENYL)ACETAMIDE | | Authors: | Lee, J, Choi, H, Kim, K.H, Jeong, S, Park, J.W, Baek, C.S, Lee, S.H. | | Deposit date: | 2007-09-05 | | Release date: | 2008-09-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Synthesis and biological evaluation of 3,5-diaminoindazoles as cyclin-dependent kinase inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
1CAD
 
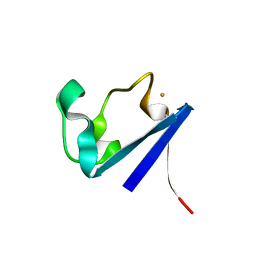 | | X-RAY CRYSTAL STRUCTURES OF THE OXIDIZED AND REDUCED FORMS OF THE RUBREDOXIN FROM THE MARINE HYPERTHERMOPHILIC ARCHAEBACTERIUM PYROCOCCUS FURIOSUS | | Descriptor: | FE (III) ION, RUBREDOXIN | | Authors: | Day, M.W, Hsu, B.T, Joshua-Tor, L, Park, J.B, Zhou, Z.H, Adams, M.W.W, Rees, D.C. | | Deposit date: | 1992-05-18 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structures of the oxidized and reduced forms of the rubredoxin from the marine hyperthermophilic archaebacterium Pyrococcus furiosus.
Protein Sci., 1, 1992
|
|
7C4R
 
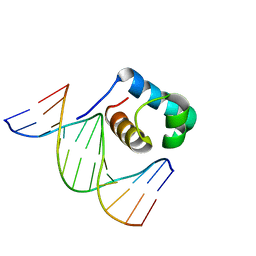 | | Crystal structure of hydrogen peroxide treated zebrafish TRF2 complexed with DNA | | Descriptor: | DNA (5'-D(*D*CP*DP*CP*DP*CP*DP*TP*DP*AP*DP*AP*DP*CP*DP*CP*DP*CP*DP*TP*DP*AP*DP*A)-3'), DNA (5'-D(*D*TP*DP*TP*DP*AP*DP*GP*DP*GP*DP*GP*DP*TP*DP*TP*DP*AP*DP*G)-3'), DNA (5'-D(*D*TP*DP*TP*DP*AP*DP*GP*DP*GP*DP*GP*DP*TP*DP*TP*DP*AP*DP*GP*DP*GP*DP*G)-3'), ... | | Authors: | Jin, Z, Park, J.H, Yun, J.H, Park, S.Y, Lee, W. | | Deposit date: | 2020-05-18 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal structure of hydrogen peroxide treated zebrafish TRF2 myb-domain complexed with DNA
To Be Published
|
|
1CAA
 
 | | X-RAY CRYSTAL STRUCTURES OF THE OXIDIZED AND REDUCED FORMS OF THE RUBREDOXIN FROM THE MARINE HYPERTHERMOPHILIC ARCHAEBACTERIUM PYROCOCCUS FURIOSUS | | Descriptor: | FE (III) ION, RUBREDOXIN | | Authors: | Day, M.W, Hsu, B.T, Joshua-Tor, L, Park, J.B, Zhou, Z.H, Adams, M.W.W, Rees, D.C. | | Deposit date: | 1992-05-18 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structures of the oxidized and reduced forms of the rubredoxin from the marine hyperthermophilic archaebacterium Pyrococcus furiosus.
Protein Sci., 1, 1992
|
|
5Y7D
 
 | | Crystal structure of human Endothelial-overexpressed LPS associated factor 1 | | Descriptor: | CHLORIDE ION, GLYCEROL, Protein CXorf40A, ... | | Authors: | Park, S.H, Kim, M.J, Park, J.S, Kim, H.J, Han, B.W. | | Deposit date: | 2017-08-17 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal Structure of Human EOLA1 Implies Its Possibility of RNA Binding.
Molecules, 24, 2019
|
|
5ZTK
 
 | | Synchrotron structure of light-driven chloride pump having an NTQ motif | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Park, J.H, Park, S.Y, Lee, W. | | Deposit date: | 2018-05-04 | | Release date: | 2018-12-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Non-cryogenic structure of a chloride pump provides crucial clues to temperature-dependent channel transport efficiency
J. Biol. Chem., 294, 2019
|
|
6AB9
 
 | | The crystal structure of the relaxed state of Nonlabens marinus Rhodopsin 3 | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.-H, Ohki, M, Park, J.-H, Jin, Z, Lee, W, Liu, H, Tame, J.R.H, Shibayama, N, Park, S.-Y. | | Deposit date: | 2018-07-20 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The pumping mechanism of NM-R3, a light-driven cyanobacterial chloride importer in the rhodopsin family
To Be Published
|
|
6ABA
 
 | | The crystal structure of the photoactivated state of Nonlabens marinus Rhodopsin 3 | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.-H, Ohki, M, Park, J.-H, Jin, Z, Lee, W, Liu, H, Tame, J.R.H, Shibayama, N, Park, S.-Y. | | Deposit date: | 2018-07-20 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | The pumping mechanism of NM-R3, a light-driven marine bacterial chloride importer in the rhodopsin family
To Be Published
|
|
5BP0
 
 | | X-ray crystal structure of Lymnaea stagnalis acetylcholine binding protein (Ls-AChBP) in complex with 5-Fluoronicotine (TI-4650) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-fluoronicotine, Acetylcholine-binding protein, ... | | Authors: | Bobango, J, Sankaran, B, Park, J.F, Wu, J, Talley, T.T. | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Comparisons of Binding Affinities for Neuronal Nicotinic Receptors (NNRs) and AChBPs
To Be Published
|
|
5BRX
 
 | | X-ray crystal structure of Aplysia californica (Ac-AChBP) in complex with 2-pyridyl azatricyclo[3.3.1.13,7]decane; 2-pyridylazaadamantane; 2-Aza (TI-8480) | | Descriptor: | (2R,3S,5R,7S)-2-(pyridin-3-yl)-1-azatricyclo[3.3.1.1~3,7~]decane, SULFATE ION, Soluble acetylcholine receptor, ... | | Authors: | Bobango, J, Sankaran, B, Park, J.F, Wu, J, Talley, T.T. | | Deposit date: | 2015-06-01 | | Release date: | 2015-06-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Comparisons of Binding Affinities for Neuronal Nicotinic Receptors (NNRs) and AChBPs
To Be Published
|
|
8IB8
 
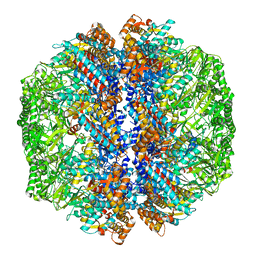 | | Human TRiC-PhLP2A-actin complex in the closed state | | Descriptor: | ACTB protein (Fragment), Phosducin-like protein 3, T-complex protein 1 subunit alpha, ... | | Authors: | Roh, S.H, Park, J, Kim, H, Lim, S. | | Deposit date: | 2023-02-09 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.42 Å) | | Cite: | A structural vista of phosducin-like PhLP2A-chaperonin TRiC cooperation during the ATP-driven folding cycle.
Nat Commun, 15, 2024
|
|
8G82
 
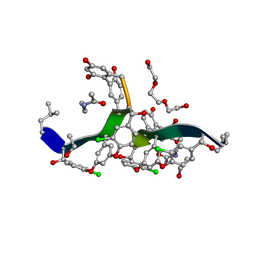 | | Vancomycin bound to D-Ala-D-Ser | | Descriptor: | BORIC ACID, D-Ala-D-Ser, DIMETHYL SULFOXIDE, ... | | Authors: | Loll, P.J, Park, J.H. | | Deposit date: | 2023-02-17 | | Release date: | 2024-01-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of vancomycin bound to the resistance determinant D-alanine-D-serine.
Iucrj, 11, 2024
|
|
8I9U
 
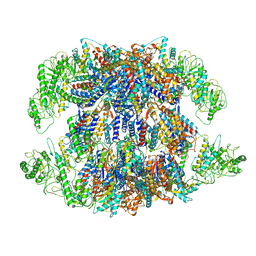 | | Human TRiC-PhLP2A complex in the open state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Phosducin-like protein 3, T-complex protein 1 subunit alpha, ... | | Authors: | Roh, S.H, Park, J, Kim, H, Lim, S. | | Deposit date: | 2023-02-07 | | Release date: | 2024-01-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A structural vista of phosducin-like PhLP2A-chaperonin TRiC cooperation during the ATP-driven folding cycle.
Nat Commun, 15, 2024
|
|
8I1U
 
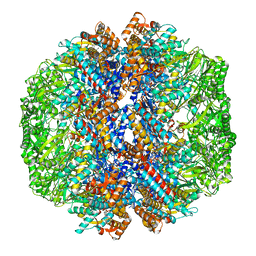 | | Human TRiC-PhLP2A complex in the closed state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Roh, S.H, Park, J, Kim, H, Lim, S. | | Deposit date: | 2023-01-13 | | Release date: | 2024-01-31 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | A structural vista of phosducin-like PhLP2A-chaperonin TRiC cooperation during the ATP-driven folding cycle.
Nat Commun, 15, 2024
|
|
8IEC
 
 | | Cryo-EM structure of miniGo-scFv16 of GPR156-miniGo-scFv16 complex (local refine) | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(o) subunit alpha, ... | | Authors: | Shin, J, Park, J, Cho, Y. | | Deposit date: | 2023-02-15 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Constitutive activation mechanism of a class C GPCR.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8IED
 
 | | Cryo-EM structure of GPR156-miniGo-scFv16 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(o) subunit alpha, ... | | Authors: | Shin, J, Park, J, Cho, Y. | | Deposit date: | 2023-02-15 | | Release date: | 2024-02-14 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Constitutive activation mechanism of a class C GPCR.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8IEB
 
 | |
8IEP
 
 | |
8IEQ
 
 | | Cryo-EM structure of G-protein free GPR156 | | Descriptor: | Probable G-protein coupled receptor 156, [(2R)-3-[(E)-hexadec-9-enoyl]oxy-2-octadecanoyloxy-propyl] 2-(trimethylazaniumyl)ethyl phosphate | | Authors: | Shin, J, Park, J, Cho, Y. | | Deposit date: | 2023-02-15 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Constitutive activation mechanism of a class C GPCR.
Nat.Struct.Mol.Biol., 31, 2024
|
|
