7YSI
 
 | | Crystal structure of thioredoxin 2 | | Descriptor: | Thiol disulfide reductase thioredoxin, ZINC ION | | Authors: | Chang, Y.J, Park, H.H. | | Deposit date: | 2022-08-12 | | Release date: | 2023-03-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.202 Å) | | Cite: | Comparison of the structure and activity of thioredoxin 2 and thioredoxin 1 from Acinetobacter baumannii.
Iucrj, 10, 2023
|
|
4UZ0
 
 | | Crystal Structure of apoptosis repressor with CARD (ARC) | | Descriptor: | GLYCEROL, NUCLEOLAR PROTEIN 3 | | Authors: | Kim, S.H, Jeong, J.H, Jang, T.H, Kim, Y.G, Park, H.H. | | Deposit date: | 2014-09-04 | | Release date: | 2015-07-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Crystal Structure of Caspase Recruiting Domain (Card) of Apoptosis Repressor with Card (Arc) and its Implication in Inhibition of Apoptosis.
Sci.Rep., 5, 2015
|
|
7BXZ
 
 | |
8WEQ
 
 | | p-hydroxybenzoate 3-monooxygenase | | Descriptor: | 4-hydroxybenzoate 3-monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Kim, S.B, Park, H.H. | | Deposit date: | 2023-09-18 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Structure of p-hydroxybenzoate 3-monooxygenase
To Be Published
|
|
8WRX
 
 | | Anti-CRISPR type II-A 28 | | Descriptor: | Anti-CRISPR protein AcrIIA28, ZINC ION | | Authors: | Kim, G.E, Park, H.H. | | Deposit date: | 2023-10-16 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | AcrIIA28 is a metalloprotein that specifically inhibits targeted-DNA loading to SpyCas9 by binding to the REC3 domain.
Nucleic Acids Res., 52, 2024
|
|
4D2K
 
 | |
8IWL
 
 | | A.baumannii Uncharacterized sugar kinase ydjH | | Descriptor: | Uncharacterized sugar kinase YdjH | | Authors: | Lee, G.H, Park, H.H. | | Deposit date: | 2023-03-30 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structure of YdjH from Acinetobacter baumannii revealed an active site of YdjH family sugar kinase.
Biochem.Biophys.Res.Commun., 664, 2023
|
|
5EZ5
 
 | |
7VI8
 
 | | Crystal structure of ChbG | | Descriptor: | ACETATE ION, Chitooligosaccharide deacetylase, ZINC ION | | Authors: | Lee, S.Y, Park, H.H. | | Deposit date: | 2021-09-26 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of ChbG from Klebsiella pneumoniae reveals the molecular basis of diacetylchitobiose deacetylation.
Commun Biol, 5, 2022
|
|
7XAO
 
 | | Crystal structure of thioredoxin 1 | | Descriptor: | Thioredoxin | | Authors: | Chang, Y.J, Park, H.H. | | Deposit date: | 2022-03-18 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High-resolution crystal structure of Acinetobacter baumannii thioredoxin 1.
Biochem.Biophys.Res.Commun., 608, 2022
|
|
2Z3B
 
 | | Crystal Structure of Bacillus Subtilis CodW, a non-canonical HslV-like peptidase with an impaired catalytic apparatus | | Descriptor: | ATP-dependent protease hslV, SODIUM ION | | Authors: | Rho, S.H, Park, H.H, Kang, G.B, Lim, Y.J, Kang, M.S, Lim, B.K, Seong, I.S, Chung, C.H, Wang, J, Eom, S.H. | | Deposit date: | 2007-06-03 | | Release date: | 2008-03-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Bacillus subtilis CodW, a noncanonical HslV-like peptidase with an impaired catalytic apparatus
Proteins, 71, 2007
|
|
2Z3A
 
 | | Crystal Structure of Bacillus Subtilis CodW, a non-canonical HslV-like peptidase with an impaired catalytic apparatus | | Descriptor: | ATP-dependent protease hslV | | Authors: | Rho, S.H, Park, H.H, Kang, G.B, Lim, Y.J, Kang, M.S, Lim, B.K, Seong, I.S, Chung, C.H, Wang, J, Eom, S.H. | | Deposit date: | 2007-06-03 | | Release date: | 2008-03-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of Bacillus subtilis CodW, a noncanonical HslV-like peptidase with an impaired catalytic apparatus
Proteins, 71, 2007
|
|
5H10
 
 | | TRAF1-TANk complex | | Descriptor: | TNF receptor-associated factor 1, TRAF family member-associated NF-kappaB activator | | Authors: | Kim, C.M, Park, H.H. | | Deposit date: | 2016-10-07 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.205 Å) | | Cite: | TRAF1-TANk complex
To Be Published
|
|
7V6E
 
 | | DREP3 | | Descriptor: | DNAation factor-related protein 3, isoform A | | Authors: | Lee, S.Y, Park, H.H. | | Deposit date: | 2021-08-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Helical filament structure of the DREP3 CIDE domain reveals a unified mechanism of CIDE-domain assembly.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
1YYF
 
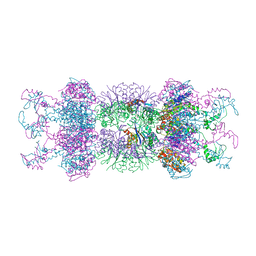 | | Correction of X-ray Intensities from an HslV-HslU co-crystal containing lattice translocation defects | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent hsl protease ATP-binding subunit hslU, ATP-dependent protease hslV | | Authors: | Wang, J, Rho, S.H, Park, H.H, Eom, S.H. | | Deposit date: | 2005-02-24 | | Release date: | 2005-07-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (4.16 Å) | | Cite: | Correction of X-ray intensities from an HslV-HslU co-crystal containing lattice-translocation defects.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
6L8P
 
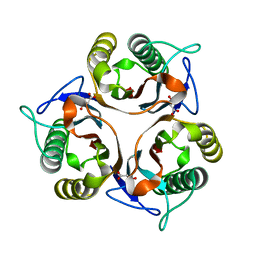 | | Crystal structure of RidA from Antarctic bacterium Psychrobacter sp. PAMC 21119 | | Descriptor: | MALONATE ION, RidA family protein | | Authors: | Kwon, S, Lee, C.W, Koh, H.Y, Lee, J.H, Park, H.H. | | Deposit date: | 2019-11-06 | | Release date: | 2019-12-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Crystal structure of the reactive intermediate/imine deaminase A homolog from the Antarctic bacterium Psychrobacter sp. PAMC 21119.
Biochem.Biophys.Res.Commun., 522, 2020
|
|
7EZY
 
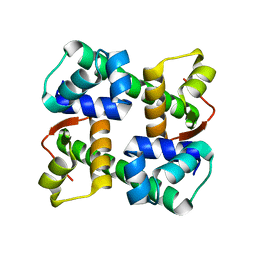 | | anti-CRISPR-associated Aca2 | | Descriptor: | anti-CRISPR-associated Aca2 | | Authors: | Lee, S.Y, Park, H.H. | | Deposit date: | 2021-06-02 | | Release date: | 2022-01-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Molecular basis of transcriptional repression ofanti-CRISPR by anti-CRISPR-associated 2
Acta Crystallogr.,Sect.D, 78, 2022
|
|
7F7P
 
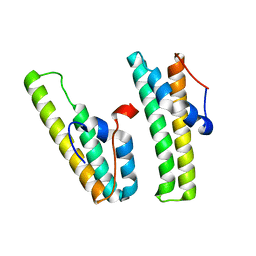 | | AcrIIC4 | | Descriptor: | anti-CRISPR protein AcrIIC4 | | Authors: | Kim, G.E, Park, H.H. | | Deposit date: | 2021-06-30 | | Release date: | 2022-05-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of the anti-CRISPR, AcrIIC4.
Protein Sci., 30, 2021
|
|
7CHR
 
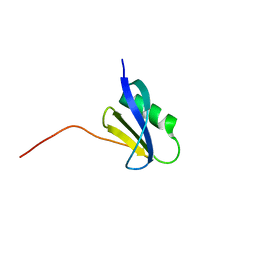 | | AcrIF9 | | Descriptor: | anti-CRISPR AcrIF9 | | Authors: | Kim, G.E, Park, H.H. | | Deposit date: | 2020-07-06 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | A high-resolution (1.2 angstrom ) crystal structure of the anti-CRISPR protein AcrIF9.
Febs Open Bio, 10, 2020
|
|
7C3K
 
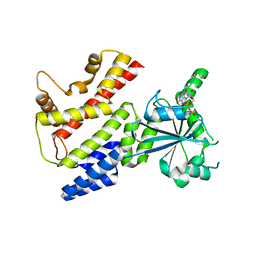 | | Crystal Structure of mIRGB10 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Immunity-related GTPase family member b10 | | Authors: | Ha, H.J, Jeong, J.H, Kim, Y.G, Park, H.H. | | Deposit date: | 2020-05-12 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular basis of IRGB10 oligomerization and membrane association for pathogen membrane disruption.
Commun Biol, 4, 2021
|
|
7CHQ
 
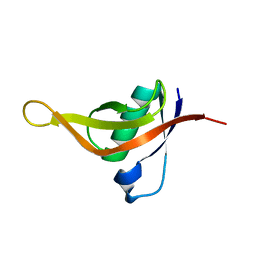 | | AcrIE2 | | Descriptor: | anti-CRISPR AcrIE2 | | Authors: | Lee, S.Y, Park, H.H. | | Deposit date: | 2020-07-06 | | Release date: | 2021-05-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | A 1.3 angstrom high-resolution crystal structure of an anti-CRISPR protein, AcrI E2.
Biochem.Biophys.Res.Commun., 533, 2020
|
|
7CP1
 
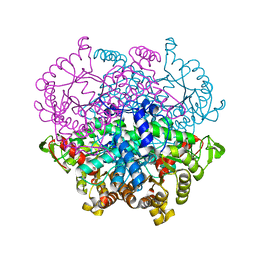 | |
7ESJ
 
 | |
