1T2K
 
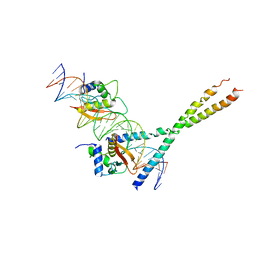 | | Structure Of The DNA Binding Domains Of IRF3, ATF-2 and Jun Bound To DNA | | Descriptor: | 31-MER, Cyclic-AMP-dependent transcription factor ATF-2, Interferon regulatory factor 3, ... | | Authors: | Panne, D, Maniatis, T, Harrison, S.C. | | Deposit date: | 2004-04-21 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of ATF-2/c-Jun and IRF-3 bound to the interferon-beta enhancer.
Embo J., 23, 2004
|
|
2O6G
 
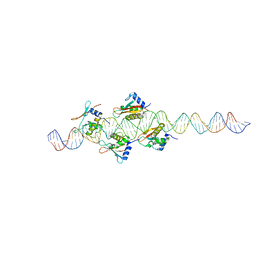 | |
2O61
 
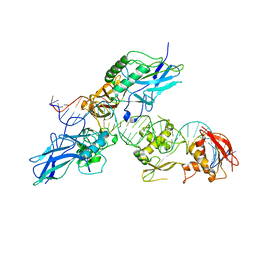 | |
6RSR
 
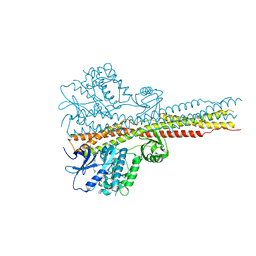 | | TBK1 in complex with compound 2 | | Descriptor: | Serine/threonine-protein kinase TBK1, ~{N}-(cyclopropen-1-ylmethyl)-2-[[4-[[4-[3,3,3-tris(fluoranyl)propanoyl]piperazin-1-yl]methyl]pyridin-2-yl]amino]-1~{H}-benzimidazole-5-carboxamide | | Authors: | Panne, D, Hillig, R.C, Rengachari, S. | | Deposit date: | 2019-05-22 | | Release date: | 2020-01-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Discovery of BAY-985, a Highly Selective TBK1/IKK epsilon Inhibitor.
J.Med.Chem., 63, 2020
|
|
6RST
 
 | | TBK1 in complex with inhibitor compound 24 | | Descriptor: | 1-[4-[(1~{R})-1-[2-[[5-[1-(cyclopropylmethyl)pyrazol-4-yl]-1~{H}-benzimidazol-2-yl]amino]pyridin-4-yl]ethyl]piperazin-1-yl]-3,3,3-tris(fluoranyl)propan-1-one, Serine/threonine-protein kinase TBK1 | | Authors: | Panne, D, Hillig, R.C, Rengachari, S. | | Deposit date: | 2019-05-22 | | Release date: | 2020-01-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Discovery of BAY-985, a Highly Selective TBK1/IKK epsilon Inhibitor.
J.Med.Chem., 63, 2020
|
|
6QPW
 
 | | Structural basis of cohesin ring opening | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Sister chromatid cohesion protein 1, ... | | Authors: | Panne, D, Muir, K.W, Li, Y, Weis, F. | | Deposit date: | 2019-02-15 | | Release date: | 2020-02-05 | | Last modified: | 2020-03-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The structure of the cohesin ATPase elucidates the mechanism of SMC-kleisin ring opening.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6RSU
 
 | | TBK1 in complex with Inhibitor compound 35 | | Descriptor: | 3,3,3-tris(fluoranyl)-1-[4-[(1~{R})-1-[2-[[(2~{S})-5-(5-propan-2-yloxypyrimidin-4-yl)-2,3-dihydro-1~{H}-benzimidazol-2-yl]amino]pyridin-4-yl]ethyl]piperazin-1-yl]propan-1-one, Serine/threonine-protein kinase TBK1 | | Authors: | Panne, D, Hillig, R.C, Rengachari, S. | | Deposit date: | 2019-05-22 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Discovery of BAY-985, a Highly Selective TBK1/IKK epsilon Inhibitor.
J.Med.Chem., 63, 2020
|
|
6GYT
 
 | |
6GYR
 
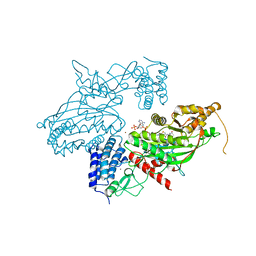 | | Transcription factor dimerization activates the p300 acetyltransferase | | Descriptor: | Histone acetyltransferase p300, ZINC ION, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]methyl (3R,20R)-20-carbamoyl-3-hydroxy-2,2-dimethyl-4,8,14,22-tetraoxo-12-thia-5,9,15,21-tetraazatricos-1-yl dihydrogen diphosphate | | Authors: | Panne, D, Ortega, E. | | Deposit date: | 2018-07-01 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Transcription factor dimerization activates the p300 acetyltransferase.
Nature, 562, 2018
|
|
4IWQ
 
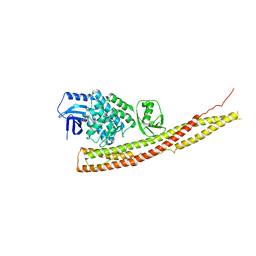 | | Crystal structure and mechanism of activation of TBK1 | | Descriptor: | N-{3-[(5-cyclopropyl-2-{[3-(morpholin-4-ylmethyl)phenyl]amino}pyrimidin-4-yl)amino]propyl}cyclobutanecarboxamide, Serine/threonine-protein kinase TBK1 | | Authors: | Panne, D, Larabi, A. | | Deposit date: | 2013-01-24 | | Release date: | 2013-03-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure and mechanism of activation of TANK-binding kinase 1.
Cell Rep, 3, 2013
|
|
4IWO
 
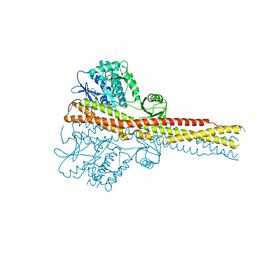 | | Crystal structure and mechanism of activation of TBK1 | | Descriptor: | N-{3-[(5-cyclopropyl-2-{[3-(2-oxopyrrolidin-1-yl)phenyl]amino}pyrimidin-4-yl)amino]propyl}cyclobutanecarboxamide, Serine/threonine-protein kinase TBK1 | | Authors: | Panne, D, Larabi, A. | | Deposit date: | 2013-01-24 | | Release date: | 2013-03-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure and mechanism of activation of TANK-binding kinase 1.
Cell Rep, 3, 2013
|
|
4IWP
 
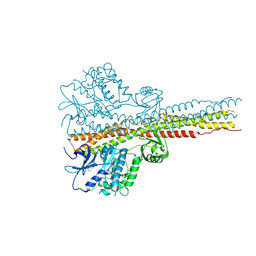 | | Crystal structure and mechanism of activation of TBK1 | | Descriptor: | N-(3-{[5-iodo-4-({3-[(thiophen-2-ylcarbonyl)amino]propyl}amino)pyrimidin-2-yl]amino}phenyl)pyrrolidine-1-carboxamide, Serine/threonine-protein kinase TBK1 | | Authors: | Panne, D, Larabi, A. | | Deposit date: | 2013-01-24 | | Release date: | 2013-03-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.065 Å) | | Cite: | Crystal structure and mechanism of activation of TANK-binding kinase 1.
Cell Rep, 3, 2013
|
|
7ZJS
 
 | |
5LKX
 
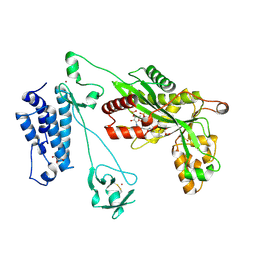 | | Crystal structure of the p300 acetyltransferase catalytic core with propionyl-coenzyme A. | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, Histone acetyltransferase p300,Histone acetyltransferase p300, ... | | Authors: | Kaczmarska, Z, Ortega, E, Marquez, J.A, Panne, D. | | Deposit date: | 2016-07-25 | | Release date: | 2016-11-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structure of p300 in complex with acyl-CoA variants.
Nat. Chem. Biol., 13, 2017
|
|
6H8Q
 
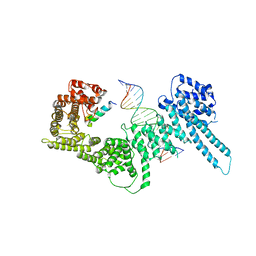 | | Structural basis for Scc3-dependent cohesin recruitment to chromatin | | Descriptor: | Cohesin subunit SCC3, DNA (5'-D(P*CP*TP*TP*TP*CP*GP*TP*TP*TP*CP*CP*TP*TP*GP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*CP*AP*AP*GP*GP*AP*AP*AP*CP*GP*AP*AP*AP*G)-3'), ... | | Authors: | Li, Y, Muir, K, Panne, D. | | Deposit date: | 2018-08-03 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.631 Å) | | Cite: | Structural basis for Scc3-dependent cohesin recruitment to chromatin.
Elife, 7, 2018
|
|
3FDQ
 
 | |
4BHW
 
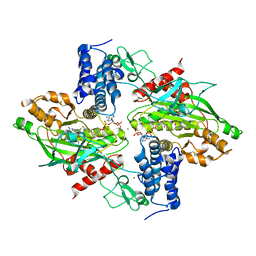 | | Structural basis for autoinhibition of the acetyltransferase activity of p300 | | Descriptor: | HISTONE ACETYLTRANSFERASE P300, ZINC ION, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]methyl (3R,20R)-20-carbamoyl-3-hydroxy-2,2-dimethyl-4,8,14,22-tetraoxo-12-thia-5,9,15,21-tetraazatricos-1-yl dihydrogen diphosphate | | Authors: | Delvecchio, M, Gaucher, J, Aguilar-Gurrieri, C, Ortega, E, Panne, D. | | Deposit date: | 2013-04-08 | | Release date: | 2013-08-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Structure of the P300 Catalytic Core and Implications for Chromatin Targeting and Hat Regulation
Nat.Struct.Mol.Biol., 20, 2013
|
|
6QPQ
 
 | |
6QNX
 
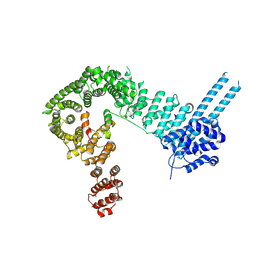 | | Structure of the SA2/SCC1/CTCF complex | | Descriptor: | 64-kDa C-terminal product, Cohesin subunit SA-2, Transcriptional repressor CTCF | | Authors: | Li, Y, Muir, K.W, Panne, D. | | Deposit date: | 2019-02-12 | | Release date: | 2020-01-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structural basis for cohesin-CTCF-anchored loops.
Nature, 578, 2020
|
|
5FRR
 
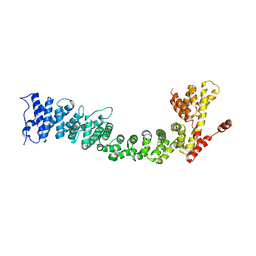 | | Structure of the Pds5-Scc1 complex and implications for cohesin function | | Descriptor: | SISTER CHROMATID COHESION PROTEIN PDS5 | | Authors: | Muir, K.W, Kschonsak, M, Li, Y, Metz, J, Haering, C.H, Panne, D. | | Deposit date: | 2015-12-22 | | Release date: | 2016-03-02 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (5.8 Å) | | Cite: | Structure of the Pds5-Scc1 Complex and Implications for Cohesin Function
Cell Rep., 14, 2016
|
|
5FRS
 
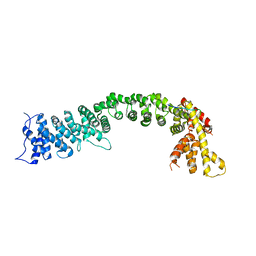 | | Structure of the Pds5-Scc1 complex and implications for cohesin function | | Descriptor: | SISTER CHROMATID COHESION PROTEIN 1, SISTER CHROMATID COHESION PROTEIN PDS5 | | Authors: | Muir, K.W, Kschonsak, M, Li, Y, Metz, J, Haering, C.H, Panne, D. | | Deposit date: | 2015-12-22 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4.073 Å) | | Cite: | Structure of the Pds5-Scc1 Complex and Implications for Cohesin Function
Cell Rep., 14, 2016
|
|
5FRP
 
 | | Structure of the Pds5-Scc1 complex and implications for cohesin function | | Descriptor: | MCD1-LIKE PROTEIN, SISTER CHROMATID COHESION PROTEIN PDS5 | | Authors: | Muir, K.W, Kschonsak, M, Li, Y, Metz, J, Haering, C.H, Panne, D. | | Deposit date: | 2015-12-21 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.895 Å) | | Cite: | Structure of the Pds5-Scc1 Complex and Implications for Cohesin Function
Cell Rep., 14, 2016
|
|
5G2E
 
 | | Structure of the Nap1 H2A H2B complex | | Descriptor: | HISTONE H2A TYPE 1, HISTONE H2B 1.1, NUCLEOSOME ASSEMBLY PROTEIN | | Authors: | AguilarGurrieri, C, Larabi, A, Vinayachandran, V, Patel, N.A, Yen, K, Reja, R, Ebong, I.O, Schoehn, G, Robinson, C.V, Pugh, B.F, Panne, D. | | Deposit date: | 2016-04-07 | | Release date: | 2016-08-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (6.7 Å) | | Cite: | Structural Evidence for Nap1-Dependent H2A-H2B Deposition and Nucleosome Assembly.
Embo J., 35, 2016
|
|
4IW0
 
 | | Crystal structure and mechanism of activation of TBK1 | | Descriptor: | N-(3-{[5-iodo-4-({3-[(thiophen-2-ylcarbonyl)amino]propyl}amino)pyrimidin-2-yl]amino}phenyl)pyrrolidine-1-carboxamide, Serine/threonine-protein kinase TBK1 | | Authors: | Larabi, A, Devos, J.M, Ng, S.-L, Nanao, M.H, Round, A, Maniatis, T, Panne, D. | | Deposit date: | 2013-01-23 | | Release date: | 2013-03-13 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystal structure and mechanism of activation of TANK-binding kinase 1.
Cell Rep, 3, 2013
|
|
5LKU
 
 | | Crystal structure of the p300 acetyltransferase catalytic core with coenzyme A. | | Descriptor: | COENZYME A, Histone acetyltransferase p300,Histone acetyltransferase p300, ZINC ION | | Authors: | Kaczmarska, Z, Ortega, E, Marquez, J.A, Panne, D. | | Deposit date: | 2016-07-25 | | Release date: | 2016-11-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of p300 in complex with acyl-CoA variants.
Nat. Chem. Biol., 13, 2017
|
|
