8FAX
 
 | | Fab 1249A8-MERS Stem Helix Complex | | Descriptor: | 1249A8-HC, 1249A8-LC, CHLORIDE ION, ... | | Authors: | Deshpande, A, Schormann, N, Piepenbrink, M.S, Martinez-Sobrido, L, Kobie, J.J, Walter, M.R. | | Deposit date: | 2022-11-28 | | Release date: | 2023-05-03 | | Last modified: | 2023-07-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and epitope of a neutralizing monoclonal antibody that targets the stem helix of beta coronaviruses.
Febs J., 290, 2023
|
|
3C8Y
 
 | | 1.39 Angstrom crystal structure of Fe-only hydrogenase | | Descriptor: | 2 IRON/2 SULFUR/3 CARBONYL/2 CYANIDE/WATER/METHYLETHER CLUSTER, FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, ... | | Authors: | Pandey, A.S, Lemon, B.J, Peters, J.W. | | Deposit date: | 2008-02-14 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Dithiomethylether as a ligand in the hydrogenase h-cluster.
J.Am.Chem.Soc., 130, 2008
|
|
2I9F
 
 | | Structure of the equine arterivirus nucleocapsid protein | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Deshpande, A, Wang, S, Walsh, M, Dokland, T. | | Deposit date: | 2006-09-05 | | Release date: | 2007-05-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the equine arteritis virus nucleocapsid protein reveals a dimer-dimer arrangement.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
1YKX
 
 | | Effect of alcohols on protein hydration | | Descriptor: | CHLORIDE ION, ETHANOL, Lysozyme C, ... | | Authors: | Deshpande, A, Nimsadkar, S, Mande, S.C. | | Deposit date: | 2005-01-18 | | Release date: | 2005-07-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Effect of alcohols on protein hydration: crystallographic analysis of hen egg-white lysozyme in the presence of alcohols.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1YKY
 
 | | Effect of alcohols on protein hydration | | Descriptor: | 1-BUTANOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Deshpande, A, Nimsadkar, S, Mande, S.C. | | Deposit date: | 2005-01-18 | | Release date: | 2005-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Effect of alcohols on protein hydration: crystallographic analysis of hen egg-white lysozyme in the presence of alcohols.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1YKZ
 
 | | Effect of alcohols on protein hydration | | Descriptor: | CHLORIDE ION, Lysozyme C, PENTANAL, ... | | Authors: | Deshpande, A, Nimsadkar, S, Mande, S.C. | | Deposit date: | 2005-01-18 | | Release date: | 2005-07-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Effect of alcohols on protein hydration: crystallographic analysis of hen egg-white lysozyme in the presence of alcohols.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1YL0
 
 | | Effect of alcohols on protein hydration | | Descriptor: | CHLORIDE ION, ISOPROPYL ALCOHOL, Lysozyme C, ... | | Authors: | Deshpande, A, Nimsadkar, S, Mande, S.C. | | Deposit date: | 2005-01-18 | | Release date: | 2005-07-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Effect of alcohols on protein hydration: crystallographic analysis of hen egg-white lysozyme in the presence of alcohols.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1Z55
 
 | | Effect of alcohols on protein hydration | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Deshpande, A, Nimsadkar, S, Mande, S.C. | | Deposit date: | 2005-03-17 | | Release date: | 2005-07-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Effect of alcohols on protein hydration: crystallographic analysis of hen egg-white lysozyme in the presence of alcohols.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1YL1
 
 | | Effect of alcohols on protein hydration | | Descriptor: | Lysozyme C, SODIUM ION, TRIFLUOROETHANOL | | Authors: | Deshpande, A.A, Nimsadkar, S, Mande, S.C. | | Deposit date: | 2005-01-18 | | Release date: | 2005-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Effect of alcohols on protein hydration: crystallographic analysis of hen egg-white lysozyme in the presence of alcohols.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2C3D
 
 | | 2.15 Angstrom crystal structure of 2-ketopropyl coenzyme M oxidoreductase carboxylase with a coenzyme M disulfide bound at the active site | | Descriptor: | 1-THIOETHANESULFONIC ACID, 2-OXOPROPYL-COM REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Pandey, A.S, Nocek, B, Clark, D.D, Ensign, S.A, Peters, J.W. | | Deposit date: | 2005-10-05 | | Release date: | 2005-11-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Mechanistic Implications of the Structure of the Mixed-Disulfide Intermediate of the Disulfide Oxidoreductase, 2-Ketopropyl-Coenzyme M Oxidoreductase/Carboxylase.
Biochemistry, 45, 2006
|
|
2C3C
 
 | | 2.01 Angstrom X-ray crystal structure of a mixed disulfide between coenzyme M and NADPH-dependent oxidoreductase 2-ketopropyl coenzyme M carboxylase | | Descriptor: | 1-THIOETHANESULFONIC ACID, 2-OXOPROPYL-COM REDUCTASE, ACETONE, ... | | Authors: | Pandey, A.S, Nocek, B, Clark, D.D, Ensign, S.A, Peters, J.W. | | Deposit date: | 2005-10-05 | | Release date: | 2005-12-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Mechanistic Implications of the Structure of the Mixed-Disulfide Intermediate of the Disulfide Oxidoreductase, 2-Ketopropyl-Coenzyme M Oxidoreductase/Carboxylase.
Biochemistry, 45, 2006
|
|
3Q6J
 
 | | Structural basis for carbon dioxide binding by 2-ketopropyl coenzyme M Oxidoreductase/Carboxylase | | Descriptor: | (2-[2-KETOPROPYLTHIO]ETHANESULFONATE, 1-THIOETHANESULFONIC ACID, 2-oxopropyl-CoM reductase, ... | | Authors: | Pandey, A.S, Mulder, D.W, Ensign, S.A, Peters, J.W. | | Deposit date: | 2011-01-01 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis for carbon dioxide binding by 2-ketopropyl coenzyme M oxidoreductase/carboxylase.
Febs Lett., 585, 2011
|
|
5I8T
 
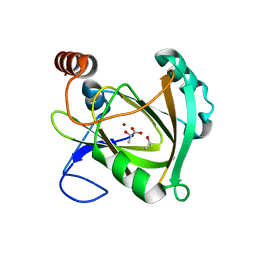 | | Structure of Mouse Acireductone dioxygenase with Ni2+ ion and D-lactic acid in the active site | | Descriptor: | 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase, ISOPROPYL ALCOHOL, LACTIC ACID, ... | | Authors: | Deshpande, A.R, Wagenpfeil, K, Pochapsky, T.C, Petsko, G.A, Ringe, D. | | Deposit date: | 2016-02-19 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Metal-Dependent Function of a Mammalian Acireductone Dioxygenase.
Biochemistry, 55, 2016
|
|
5I8Y
 
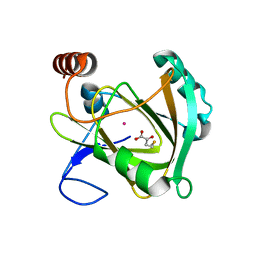 | | Structure of Mouse Acireductone Dioxygenase bound to Co2+ and 2-keto-4-(methylthio)-butyric acid | | Descriptor: | 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase, 4-(METHYLSULFANYL)-2-OXOBUTANOIC ACID, COBALT (II) ION | | Authors: | Deshpande, A.R, Wagenpfeil, K, Pochapsky, T.C, Petsko, G.A, Ringe, D. | | Deposit date: | 2016-02-19 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.942 Å) | | Cite: | Metal-Dependent Function of a Mammalian Acireductone Dioxygenase.
Biochemistry, 55, 2016
|
|
5I93
 
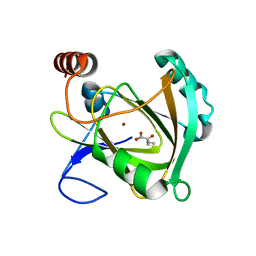 | | Structure of Mouse Acireductone dioxygenase with Ni2+ and 2-ketopentanoic acid in the active site | | Descriptor: | 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase, 2-oxopentanoic acid, NICKEL (II) ION | | Authors: | Deshpande, A.R, Wagenpfeil, K, Pochapsky, T.C, Petsko, G.A, Ringe, D. | | Deposit date: | 2016-02-19 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.236 Å) | | Cite: | Metal-Dependent Function of a Mammalian Acireductone Dioxygenase.
Biochemistry, 55, 2016
|
|
5I91
 
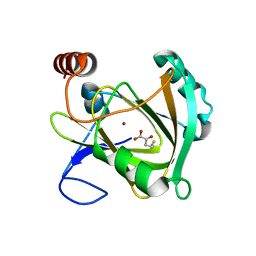 | | Structure of Mouse Acirecutone dioxygenase with to Ni2+ and 2-keto-4-(methylthio)-butyric acid in the active site | | Descriptor: | 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase, 4-(METHYLSULFANYL)-2-OXOBUTANOIC ACID, NICKEL (II) ION | | Authors: | Deshpande, A.R, Robinson, H, Wagenpfeil, K, Pochapsky, T.C, Petsko, G.A, Ringe, D. | | Deposit date: | 2016-02-19 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Metal-Dependent Function of a Mammalian Acireductone Dioxygenase.
Biochemistry, 55, 2016
|
|
5I8S
 
 | | Structure of Mouse Acireductone dioxygenase with Ni2+ ion and pentanoic acid in the active site | | Descriptor: | 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase, NICKEL (II) ION, PENTANOIC ACID | | Authors: | Deshpande, A.R, Wagenpfeil, K, Pochapsky, T.C, Petsko, G.A, Ringe, D. | | Deposit date: | 2016-02-19 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Metal-Dependent Function of a Mammalian Acireductone Dioxygenase.
Biochemistry, 55, 2016
|
|
5H88
 
 | | Crystal structure of mRojoA mutant - T16V -P63F - W143A - L163V | | Descriptor: | mRojoA fluorescent protein | | Authors: | Pandelieva, A.T, Tremblay, V, Sarvan, S, Chica, R.A, Couture, J.-F. | | Deposit date: | 2015-12-23 | | Release date: | 2016-01-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Brighter Red Fluorescent Proteins by Rational Design of Triple-Decker Motif.
Acs Chem.Biol., 11, 2016
|
|
5H89
 
 | | Crystal structure of mRojoA mutant - T16V - P63Y - W143G - L163V | | Descriptor: | mRojoA fluorescent protein | | Authors: | Pandelieva, A.T, Tremblay, V, Sarvan, S, Chica, R.A, Couture, J.-F. | | Deposit date: | 2015-12-23 | | Release date: | 2016-01-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Brighter Red Fluorescent Proteins by Rational Design of Triple-Decker Motif.
Acs Chem.Biol., 11, 2016
|
|
5H87
 
 | | Crystal structure of mRojoA mutant - P63H - W143S | | Descriptor: | mRojoA fluorescent protein | | Authors: | Pandelieva, A.T, Tremblay, V, Sarvan, S, Chica, R.A, Couture, J.-F. | | Deposit date: | 2015-12-23 | | Release date: | 2016-01-27 | | Last modified: | 2016-03-02 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Brighter Red Fluorescent Proteins by Rational Design of Triple-Decker Motif.
Acs Chem.Biol., 11, 2016
|
|
1I5I
 
 | | THE C18S MUTANT OF BOVINE (GAMMA-B)-CRYSTALLIN | | Descriptor: | (GAMMA-B) CRYSTALLIN | | Authors: | Zarutskie, J.A, Asherie, N, Pande, J, Pande, A, Lomakin, J, Lomakin, A, Ogun, O, Stern, L.J, King, J.A, Benedek, G.B. | | Deposit date: | 2001-02-27 | | Release date: | 2001-03-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Enhanced crystallization of the Cys18 to Ser mutant of bovine gammaB crystallin.
J.Mol.Biol., 314, 2001
|
|
2NBR
 
 | | The Solution Structure of Human gammaC-crystallin | | Descriptor: | Gamma-crystallin C | | Authors: | Dixit, K, Pande, A, Pande, J, Sarma, S.P. | | Deposit date: | 2016-03-12 | | Release date: | 2016-06-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Structure of a Major Lens Protein, Human gamma C-Crystallin: Role of the Dipole Moment in Protein Solubility.
Biochemistry, 55, 2016
|
|
7JXG
 
 | | Structural model for Fe-containing human acireductone dioxygenase | | Descriptor: | 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase, FE (II) ION | | Authors: | Pochapsky, T.C, Liu, X, Deshpande, A, Ringe, D, Garber, A, Ryan, J. | | Deposit date: | 2020-08-27 | | Release date: | 2020-11-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A Model for the Solution Structure of Human Fe(II)-Bound Acireductone Dioxygenase and Interactions with the Regulatory Domain of Matrix Metalloproteinase I (MMP-I).
Biochemistry, 59, 2020
|
|
1O7Q
 
 | | Roles of Individual Residues of Alpha-1,3 Galactosyltransferases in Substrate Binding and Catalysis | | Descriptor: | GLYCEROL, MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, ... | | Authors: | Zhang, Y, Swaminathan, G.J, Deshpande, A, Natesh, R, Xie, Z, Acharya, K.R, Brew, K. | | Deposit date: | 2002-11-12 | | Release date: | 2003-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Roles of Individual Enzyme-Substrate Interactions by Alpha-1,3-Galactosyltransferase in Catalysis and Specificity.
Biochemistry, 42, 2003
|
|
1O7O
 
 | | Roles of Individual Residues of Alpha-1,3 Galactosyltransferases in Substrate Binding and Catalysis | | Descriptor: | MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Zhang, Y, Swaminathan, G.J, Deshpande, A, Natesh, R, Xie, Z, Acharya, K.R, Brew, K. | | Deposit date: | 2002-11-11 | | Release date: | 2003-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Roles of individual enzyme-substrate interactions by alpha-1,3-galactosyltransferase in catalysis and specificity.
Biochemistry, 42, 2003
|
|
