6B5X
 
 | | Beta-Lactamase, unmixed shards crystal form | | Descriptor: | Beta-lactamase, PHOSPHATE ION | | Authors: | Pandey, S. | | Deposit date: | 2017-09-29 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Enzyme intermediates captured "on the fly" by mix-and-inject serial crystallography.
BMC Biol., 16, 2018
|
|
6B6F
 
 | |
4HJI
 
 | |
3OF6
 
 | | Human pre-T cell receptor crystal structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Pre T-cell antigen receptor alpha, T cell receptor beta chain | | Authors: | Pang, S.S. | | Deposit date: | 2010-08-13 | | Release date: | 2010-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structural basis for autonomous dimerization of the pre-T-cell antigen receptor
Nature, 467, 2010
|
|
3OR6
 
 | | On the structural basis of modal gating behavior in K+channels - E71Q | | Descriptor: | POTASSIUM ION, Voltage-gated potassium channel, antibody fab fragment heavy chain, ... | | Authors: | Chakrapani, S, Cordero-Morales, J.F, Jogini, V, Perozo, E. | | Deposit date: | 2010-09-06 | | Release date: | 2011-01-05 | | Last modified: | 2018-08-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | On the structural basis of modal gating behavior in K(+) channels.
Nat.Struct.Mol.Biol., 18, 2011
|
|
1II2
 
 | | Crystal Structure of Phosphoenolpyruvate Carboxykinase (PEPCK) from Trypanosoma cruzi | | Descriptor: | PHOSPHOENOLPYRUVATE CARBOXYKINASE, SULFATE ION | | Authors: | Trapani, S, Linss, J, Goldenberg, S, Fischer, H, Craievich, A.F, Oliva, G. | | Deposit date: | 2001-04-20 | | Release date: | 2001-11-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the dimeric phosphoenolpyruvate carboxykinase (PEPCK) from Trypanosoma cruzi at 2 A resolution.
J.Mol.Biol., 313, 2001
|
|
6MIC
 
 | |
7ZC9
 
 | | Human Pikachurin/EGFLAM C-terminal Laminin-G domain (LG3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Pikachurin, SULFATE ION | | Authors: | Pantalone, S, Forneris, F. | | Deposit date: | 2022-03-26 | | Release date: | 2023-07-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the photoreceptor synaptic assembly of the extracellular matrix protein pikachurin with the orphan receptor GPR179.
Sci.Signal., 16, 2023
|
|
7ZCB
 
 | | Human Pikachurin/EGFLAM N-terminal Fibronectin-III (1-2) domains | | Descriptor: | CHLORIDE ION, Pikachurin, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Pantalone, S, Savino, S, Viti, L.V, Forneris, F. | | Deposit date: | 2022-03-26 | | Release date: | 2023-07-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the photoreceptor synaptic assembly of the extracellular matrix protein pikachurin with the orphan receptor GPR179.
Sci.Signal., 16, 2023
|
|
6NQ6
 
 | | Structure & function of a new Aspartylglucosaminuria variant | | Descriptor: | N(4)-(Beta-N-acetylglucosaminyl)-L-asparaginase | | Authors: | Pande, S, Guo, H.C. | | Deposit date: | 2019-01-19 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The T99K variant of glycosylasparaginase shows a new structural mechanism of the genetic disease aspartylglucosaminuria.
Protein Sci., 28, 2019
|
|
3S0T
 
 | |
5EVF
 
 | | Crystal structure of a Francisella virulence factor FvfA in the hexagonal form | | Descriptor: | CHLORIDE ION, Francisella virulence factor, GLYCEROL | | Authors: | Kolappan, S, Lo, K.Y, Shen, C.L.J, Guttman, J.A, Craig, L. | | Deposit date: | 2015-11-19 | | Release date: | 2016-10-26 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (1.762 Å) | | Cite: | Structure of the conserved Francisella virulence protein FvfA.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5EVG
 
 | | Crystal structure of a Francisella virulence factor FvfA in the orthorhombic form | | Descriptor: | Francisella virulence factor | | Authors: | Kolappan, S, Lo, K.Y, Shen, C.L.J, Guttman, J.A, Craig, L. | | Deposit date: | 2015-11-19 | | Release date: | 2016-10-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure of the conserved Francisella virulence protein FvfA.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
6S44
 
 | |
6RTK
 
 | |
5XPJ
 
 | |
3HUJ
 
 | |
1JSC
 
 | | Crystal Structure of the Catalytic Subunit of Yeast Acetohydroxyacid Synthase: A target for Herbicidal Inhibitors | | Descriptor: | ACETOHYDROXY-ACID SYNTHASE, DIHYDROGENPHOSPHATE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Pang, S.S, Duggleby, R.G, Guddat, L.W. | | Deposit date: | 2001-08-17 | | Release date: | 2002-01-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of yeast acetohydroxyacid synthase: a target for herbicidal inhibitors.
J.Mol.Biol., 317, 2002
|
|
4YRU
 
 | | Crystal structure of C-terminally truncated Neuronal Calcium Sensor (NCS-1) from Rattus norvegicus | | Descriptor: | CALCIUM ION, Neuronal calcium sensor 1 | | Authors: | Pandalaneni, S, Karrupiah, V, Mayans, O, Derrick, J.P, Lian, L.Y. | | Deposit date: | 2015-03-15 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Neuronal Calcium Sensor-1 Binds the D2 Dopamine Receptor and G-protein-coupled Receptor Kinase 1 (GRK1) Peptides Using Different Modes of Interactions.
J.Biol.Chem., 290, 2015
|
|
7ZF2
 
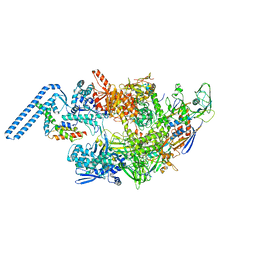 | | Protomeric substructure from an octameric assembly of M. tuberculosis RNA polymerase in complex with sigma-b initiation factor | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Trapani, S, Bron, P, Lai Kee Him, J, Brodolin, K, Morichaud, Z, Vishwakarma, R. | | Deposit date: | 2022-03-31 | | Release date: | 2023-02-08 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Structural basis of the mycobacterial stress-response RNA polymerase auto-inhibition via oligomerization.
Nat Commun, 14, 2023
|
|
3AS0
 
 | | Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - W275G mutant complex structure with Sanguinarine | | Descriptor: | 13-methyl[1,3]benzodioxolo[5,6-c][1,3]dioxolo[4,5-i]phenanthridin-13-ium, Chitinase A, GLYCEROL | | Authors: | Pantoom, S, Vetter, I.R, Prinz, H, Suginta, W. | | Deposit date: | 2010-12-09 | | Release date: | 2011-04-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Potent family-18 chitinase inhibitors: x-ray structures, affinities, and binding mechanisms
J.Biol.Chem., 286, 2011
|
|
3ARO
 
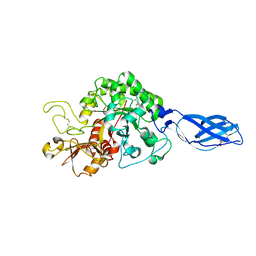 | | Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - apo structure | | Descriptor: | Chitinase A, GLYCEROL | | Authors: | Pantoom, S, Vetter, I.R, Prinz, H, Suginta, W. | | Deposit date: | 2010-12-09 | | Release date: | 2011-04-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Potent family-18 chitinase inhibitors: x-ray structures, affinities, and binding mechanisms
J.Biol.Chem., 286, 2011
|
|
3ARY
 
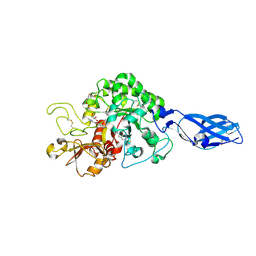 | | Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - complex structure with 2-(imidazolin-2-yl)-5-isothiocyanatobenzofuran | | Descriptor: | 2-(5-isothiocyanato-1-benzofuran-2-yl)-4,5-dihydro-1H-imidazole, Chitinase A, GLYCEROL | | Authors: | Pantoom, S, Vetter, I.R, Prinz, H, Suginta, W. | | Deposit date: | 2010-12-09 | | Release date: | 2011-04-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Potent family-18 chitinase inhibitors: x-ray structures, affinities, and binding mechanisms
J.Biol.Chem., 286, 2011
|
|
3ARS
 
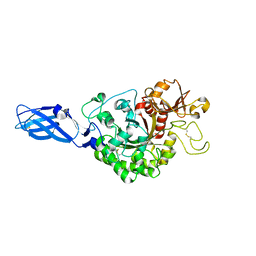 | | Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - apo structure of mutant W275G | | Descriptor: | Chitinase A | | Authors: | Pantoom, S, Vetter, I.R, Prinz, H, Suginta, W. | | Deposit date: | 2010-12-09 | | Release date: | 2011-04-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Potent family-18 chitinase inhibitors: x-ray structures, affinities, and binding mechanisms
J.Biol.Chem., 286, 2011
|
|
3ARX
 
 | | Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - complex structure with Propentofylline | | Descriptor: | 3-methyl-1-(5-oxohexyl)-7-propyl-3,7-dihydro-1H-purine-2,6-dione, Chitinase A, GLYCEROL | | Authors: | Pantoom, S, Vetter, I.R, Prinz, H, Suginta, W. | | Deposit date: | 2010-12-09 | | Release date: | 2011-04-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Potent family-18 chitinase inhibitors: x-ray structures, affinities, and binding mechanisms
J.Biol.Chem., 286, 2011
|
|
