6VU2
 
 | | M1214_N1 Fab structure | | Descriptor: | M1214 N1 Fab heavy chain, M1214 N1 Fab light chain | | Authors: | Pan, R, Kong, X. | | Deposit date: | 2020-02-14 | | Release date: | 2020-05-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | VSV-Displayed HIV-1 Envelope Identifies Broadly Neutralizing Antibodies Class-Switched to IgG and IgA.
Cell Host Microbe, 27, 2020
|
|
5UBY
 
 | | Fab structure of anti-HIV-1 gp120 mAb 1A8 | | Descriptor: | Heavy chain of Fab fragment of anti-HIV1 gp120 mAb 1A8, Light chain of Fab fragment of anti-HIV1 gp120 mAb 1A8 | | Authors: | Pan, R, Kong, X.-P. | | Deposit date: | 2016-12-21 | | Release date: | 2018-01-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | High Antibody Diversity and Low Inter-clonal Competition Favor Production of Functional Neutralizing Antibodies
To be Published
|
|
5UBZ
 
 | | Fab structure of HIV gp120 specific mAb 1E12 | | Descriptor: | Anti-HIV1 gp120 mAb 1E12 Fab heavy chain, Anti-HIV1 gp120 mAb 1E12 Fab light chain | | Authors: | Pan, R, Kong, X.-P. | | Deposit date: | 2016-12-21 | | Release date: | 2018-01-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | High Antibody Diversity and Low Inter-clonal Competition Favor Production of Functional Neutralizing Antibodies
To be published
|
|
5V6M
 
 | | Crystal Structure of Rabbit Anti-HIV-1 gp120 V3 Fab 10A3 in complex with V3 peptide ConB | | Descriptor: | CALCIUM ION, Envelope glycoprotein gp120 V3 peptide of Con B sequence, Heavy chain of Fab fragment of rabbit anti-HIV1 gp120 V3 mAb 10A3, ... | | Authors: | Pan, R, Kong, X.-P. | | Deposit date: | 2017-03-17 | | Release date: | 2018-01-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Increased epitope complexity correlated with antibody affinity maturation and a novel binding mode revealed by structures of rabbit antibodies against the third variable loop (V3) of HIV-1 gp120.
J. Virol., 2018
|
|
5V6L
 
 | | Crystal Structure of Rabbit Anti-HIV-1 gp120 V3 Fab 10A37 in complex with V3 peptide JR-FL | | Descriptor: | Envelope glycoprotein, v3 region, Heavy chain of Fab fragment of rabbit anti-HIV1 gp120 V3 mAb 10A37, ... | | Authors: | Pan, R, Kong, X.-P. | | Deposit date: | 2017-03-17 | | Release date: | 2018-01-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.549 Å) | | Cite: | Increased epitope complexity correlated with antibody affinity maturation and a novel binding mode revealed by structures of rabbit antibodies against the third variable loop (V3) of HIV-1 gp120.
J. Virol., 2018
|
|
4XML
 
 | | Crystal structure of Fab of HIV-1 gp120 V3-specific human monoclonal antibody 2424 | | Descriptor: | Heavy chain of HIV-1 gp120 V3-specific human monoclonal antibody 2424, Light chain of HIV-1 gp120 V3-specific human monoclonal antibody 2424 | | Authors: | Pan, R, Kong, X.-P. | | Deposit date: | 2015-01-14 | | Release date: | 2015-07-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Functional and Structural Characterization of Human V3-Specific Monoclonal Antibody 2424 with Neutralizing Activity against HIV-1 JRFL.
J.Virol., 89, 2015
|
|
4ZTO
 
 | |
4YWG
 
 | | Crystal structure of 830A in complex with V1V2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of anti-HIV-1 gp120 V1V2 antibody 830A, Light chain of anti-HIV-1 gp120 V1V2 antibody 830A, ... | | Authors: | Pan, R, Kong, X. | | Deposit date: | 2015-03-20 | | Release date: | 2015-07-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.998 Å) | | Cite: | The V1V2 Region of HIV-1 gp120 Forms a Five-Stranded Beta Barrel.
J.Virol., 89, 2015
|
|
4ZTP
 
 | |
9FYB
 
 | | Structural Insights into the NMN Complex of Nicotinate Nucleotide Adenylyltransferase from Enterococcus faecium via Co-Crystallization Studies | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Pandian, R, Jeje, O.A, Sayed, Y, Achilonu, I.A. | | Deposit date: | 2024-07-03 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural Insights into the NMN Complex of Nicotinate Nucleotide Adenylyltransferase from Enterococcus faecium via Co-Crystallization Studies
To be published
|
|
6Y1E
 
 | | Crystal structure of human glutathione transferase P1-1 (hGSTP1-1) that was co-crystallised in the presence of indanyloxyacetic acid-94 (IAA-94) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-[[6,7-bis(chloranyl)-2-cyclopentyl-2-methyl-1-oxidanylidene-3~{H}-inden-5-yl]oxy]ethanoic acid, GLUTATHIONE, ... | | Authors: | Pandian, R, Worth, R, Thangaraj, V, Sayed, Y, Dirr, H.W. | | Deposit date: | 2020-02-12 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.402 Å) | | Cite: | The interaction of IAA-94 with the soluble conformation of the CLIC1 protein and its structural homolog hGSTP1-1
To Be Published
|
|
6W2S
 
 | | Structure of the Cricket Paralysis Virus 5-UTR IRES (CrPV 5-UTR-IRES) bound to the small ribosomal subunit in the open state (Class 1) | | Descriptor: | 18S rRNA, CrPV 5'-UTR IRES, Eukaryotic translation initiation factor 3 subunit A, ... | | Authors: | Neupane, R, Pisareva, V, Rodriguez, C.F, Pisarev, A, Fernandez, I.S. | | Deposit date: | 2020-03-08 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | A complex IRES at the 5'-UTR of a viral mRNA assembles a functional 48S complex via an uAUG intermediate.
Elife, 9, 2020
|
|
6W2T
 
 | | Structure of the Cricket Paralysis Virus 5-UTR IRES (CrPV 5-UTR-IRES) bound to the small ribosomal subunit in the closed state (Class 2) | | Descriptor: | 18S rRNA, CrPV 5'-UTR IRES, Eukaryotic translation initiation factor 3 subunit A, ... | | Authors: | Neupane, R, Pisareva, V, Rodriguez, C.F, Pisarev, A, Fernandez, I.S. | | Deposit date: | 2020-03-08 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | A complex IRES at the 5'-UTR of a viral mRNA assembles a functional 48S complex via an uAUG intermediate.
Elife, 9, 2020
|
|
8ALS
 
 | |
8AIH
 
 | | Crystal Structure of Enterococcus faecium Nicotinate Nucleotide Adenylyltransferase at 1.9 Angstroms Resolution | | Descriptor: | DIHYDROGENPHOSPHATE ION, Probable nicotinate-nucleotide adenylyltransferase, SULFATE ION | | Authors: | Pandian, R, Jeje, O.A, Sayed, Y, Achilonu, I.A. | | Deposit date: | 2022-07-26 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Obtaining high yield recombinant Enterococcus faecium nicotinate nucleotide adenylyltransferase for X-ray crystallography and biophysical studies.
Int.J.Biol.Macromol., 250, 2023
|
|
8AII
 
 | | High Resolution Crystal Structure of Enterococcus faecium Nicotinate Nucleotide Adenylyltransferase Complexed with Adenine | | Descriptor: | ADENINE, MAGNESIUM ION, Probable nicotinate-nucleotide adenylyltransferase, ... | | Authors: | Pandian, R, Jeje, O.A, Sayed, Y, Achilonu, I.A. | | Deposit date: | 2022-07-26 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Obtaining high yield recombinant Enterococcus faecium nicotinate nucleotide adenylyltransferase for X-ray crystallography and biophysical studies.
Int.J.Biol.Macromol., 250, 2023
|
|
8BHZ
 
 | |
1BM3
 
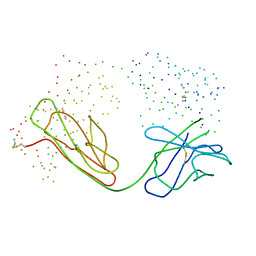 | | IMMUNOGLOBULIN OPG2 FAB-PEPTIDE COMPLEX | | Descriptor: | IMMUNOGLOBULIN OPG2 FAB, CONSTANT DOMAIN, VARIABLE DOMAIN | | Authors: | Kodandapani, R, Veerapandian, L, Ni, C.Z, Chiou, C.-K, Whital, R, Kunicki, T.J, Ely, K.R. | | Deposit date: | 1999-04-15 | | Release date: | 1999-04-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational change in an anti-integrin antibody: structure of OPG2 Fab bound to a beta 3 peptide.
Biochem.Biophys.Res.Commun., 251, 1998
|
|
1OPG
 
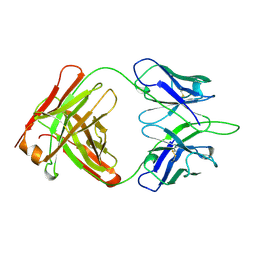 | | OPG2 FAB FRAGMENT | | Descriptor: | OPG2 FAB (HEAVY CHAIN), OPG2 FAB (LIGHT CHAIN) | | Authors: | Kodandapani, R, Veerapandian, B, Ely, K.R. | | Deposit date: | 1995-04-28 | | Release date: | 1995-07-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the OPG2 Fab. An antireceptor antibody that mimics an RGD cell adhesion site.
J.Biol.Chem., 270, 1995
|
|
6ZMX
 
 | | Crystal structure of hemoglobin from turkey (Meleagiris gallopova) crystallized in orthorhombic form at 1.4 Angstrom resolution | | Descriptor: | Hemoglobin beta chain, Hemoglobin subunit alpha-A, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Pandian, R, Shobana, N, Sundaresan, S.S, Sayed, Y, Ponnuswamy, M.N. | | Deposit date: | 2020-07-04 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.389 Å) | | Cite: | Structural studies of hemoglobin from two flightless birds, ostrich and turkey: insights into their differing oxygen-binding properties.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6ZMY
 
 | | Crystal structure of hemoglobin from turkey (Meleagiris gallopova) crystallized in monoclinic form at 1.66 Angstrom resolution | | Descriptor: | Hemoglobin beta chain, Hemoglobin subunit alpha-A, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Pandian, R, Shobana, N, Sundaresan, S.S, Thangaraj, V, Sayed, Y, Ponnuswamy, M.N. | | Deposit date: | 2020-07-04 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.655 Å) | | Cite: | Structural studies of hemoglobin from two flightless birds, ostrich and turkey: insights into their differing oxygen-binding properties.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
8IU6
 
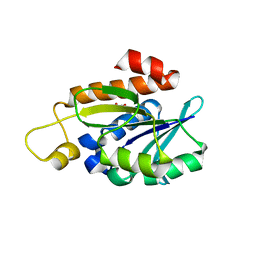 | | Crystal structure of peptidyl-tRNA hydrolase mutant from Enterococcus faecium | | Descriptor: | GLYCEROL, Peptidyl-tRNA hydrolase | | Authors: | Pandey, R, Tripathi, S, Lanka, A.K, Zohib, M, Pal, R.K, Arora, A. | | Deposit date: | 2023-03-23 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of peptidyl-tRNA hydrolase mutant from Enterococcus faecium
To Be Published
|
|
1PUE
 
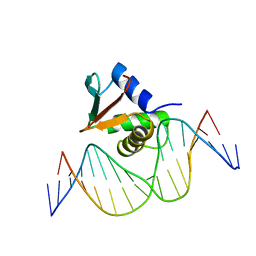 | | PU.1 ETS DOMAIN-DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*AP*GP*GP*GP*GP*AP*AP*GP*TP*GP*GP*G)-3'), DNA (5'-D(*TP*CP*CP*CP*AP*CP*TP*TP*CP*CP*CP*CP*TP*TP*TP*T)-3'), PROTEIN (TRANSCRIPTION FACTOR PU.1 (TF PU.1)) | | Authors: | Kodandapani, R, Pio, F, Ni, C.Z, Piccialli, G, Klemsz, M, McKercher, S, Maki, R.A, Ely, K.R. | | Deposit date: | 1996-07-08 | | Release date: | 1997-02-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A new pattern for helix-turn-helix recognition revealed by the PU.1 ETS-domain-DNA complex.
Nature, 380, 1996
|
|
7Y52
 
 | | Crystal structure of peptidyl-tRNA hydrolase from Enterococcus faecium | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Peptidyl-tRNA hydrolase, ... | | Authors: | Pandey, R, Zohib, M, Mundra, S, Pal, R.K, Arora, A. | | Deposit date: | 2022-06-16 | | Release date: | 2024-01-17 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Characterization of structure of peptidyl-tRNA hydrolase from Enterococcus faecium and its inhibition by a pyrrolinone compound.
Int.J.Biol.Macromol., 275, 2024
|
|
4LYM
 
 | |
