4NJR
 
 | | Structural and kinetic bases for the metal preference of the M18 aminopeptidase from Pseudomonas aeruginosa | | Descriptor: | CARBONATE ION, Probable M18 family aminopeptidase 2, ZINC ION | | Authors: | Nguyen, D.D, Pandian, R, Kim, D.Y, Ha, S.C, Yun, K.H, Kim, K.S, Kim, J.H, Kim, K.K. | | Deposit date: | 2013-11-11 | | Release date: | 2014-04-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and kinetic bases for the metal preference of the M18 aminopeptidase from Pseudomonas aeruginosa
Biochem.Biophys.Res.Commun., 447, 2014
|
|
4OID
 
 | | Structural and kinetic bases for the metal preference of the M18 aminopeptidase from Pseudomonas aeruginosa | | Descriptor: | Probable M18 family aminopeptidase 2 | | Authors: | Nguyen, D.D, Pandian, R, Kim, D.D, Ha, S.C, Yoon, H.J, Kim, K.S, Yun, K.H, Kim, J.H, Kim, K.K. | | Deposit date: | 2014-01-19 | | Release date: | 2014-04-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and kinetic bases for the metal preference of the M18 aminopeptidase from Pseudomonas aeruginosa
Biochem.Biophys.Res.Commun., 447, 2014
|
|
4OIW
 
 | | Structural and kinetic bases for the metal preference of the M18 aminopeptidase from Pseudomonas aeruginosa | | Descriptor: | Probable M18 family aminopeptidase 2, ZINC ION | | Authors: | Nguyen, D.D, Pandian, R, Kim, D.D, Ha, S.C, Yoon, H.J, Kim, K.S, Yun, K.H, Kim, J.H, Kim, K.K. | | Deposit date: | 2014-01-20 | | Release date: | 2014-04-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural and kinetic bases for the metal preference of the M18 aminopeptidase from Pseudomonas aeruginosa
Biochem.Biophys.Res.Commun., 447, 2014
|
|
4NJQ
 
 | | Structural and kinetic bases for the metal preference of the M18 aminopeptidase from Pseudomonas aeruginosa | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CARBONATE ION, COBALT (II) ION, ... | | Authors: | Nguyen, D.D, Pandian, R, Kim, D.Y, Ha, S.C, Yun, K.H, Kim, K.S, Kim, J.H, Kim, K.K. | | Deposit date: | 2013-11-11 | | Release date: | 2014-04-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Structural and kinetic bases for the metal preference of the M18 aminopeptidase from Pseudomonas aeruginosa
Biochem.Biophys.Res.Commun., 447, 2014
|
|
4OL8
 
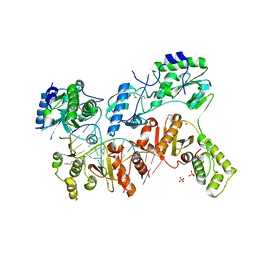 | | Ty3 reverse transcriptase bound to DNA/RNA | | Descriptor: | 5'-D(*CP*AP*TP*CP*TP*TP*CP*CP*TP*CP*TP*CP*TP*CP*TP*C)-3', 5'-R(*CP*UP*GP*AP*GP*AP*GP*AP*GP*AP*GP*GP*AP*AP*GP*AP*UP*G)-3', Reverse transcriptase/ribonuclease H, ... | | Authors: | Nowak, E, Miller, J.T, Bona, M.K, Studnicka, J, Szczepanowski, R.H, Jurkowski, J, Le Grice, S.F.J, Nowotny, M. | | Deposit date: | 2014-01-23 | | Release date: | 2014-03-05 | | Last modified: | 2014-04-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Ty3 reverse transcriptase complexed with an RNA-DNA hybrid shows structural and functional asymmetry.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4D6D
 
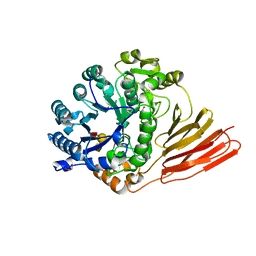 | | Crystal structure of a family 98 glycoside hydrolase catalytic module (Sp3GH98) in complex with the blood group A-trisaccharide (X02 mutant) | | Descriptor: | 1,2-ETHANEDIOL, GLYCOSIDE HYDROLASE, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose | | Authors: | Kwan, D.H, Constantinescu, I, Chapanian, R, Higgins, M.A, Samain, E, Boraston, A.B, Kizhakkedathu, J.N, Withers, S.G. | | Deposit date: | 2014-11-11 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Towards Efficient Enzymes for the Generation of Universal Blood Through Structure-Guided Directed Evolution.
J.Am.Chem.Soc., 137, 2015
|
|
4D6G
 
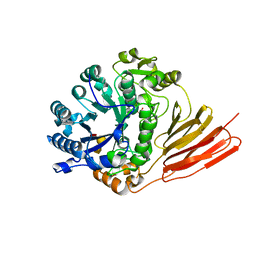 | | Crystal structure of a family 98 glycoside hydrolase catalytic module (Sp3GH98) in complex with the blood group A-trisaccharide (L19 mutant) | | Descriptor: | 1,2-ETHANEDIOL, GLYCOSIDE HYDROLASE, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]alpha-D-galactopyranose | | Authors: | Kwan, D.H, Constantinescu, I, Chapanian, R, Higgins, M.A, Samain, E, Boraston, A.B, Kizhakkedathu, J.N, Withers, S.G. | | Deposit date: | 2014-11-11 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Towards Efficient Enzymes for the Generation of Universal Blood Through Structure-Guided Directed Evolution.
J.Am.Chem.Soc., 137, 2015
|
|
4D71
 
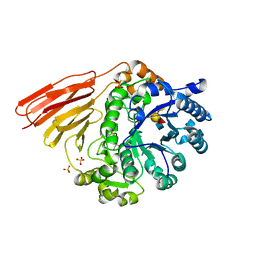 | | Crystal structure of a family 98 glycoside hydrolase catalytic module (Sp3GH98) in complex with the type 2 blood group A-tetrasaccharide (E558A X02 mutant) | | Descriptor: | 1,2-ETHANEDIOL, GLYCOSIDE HYDROLASE, SULFATE ION, ... | | Authors: | Kwan, D.H, Constantinescu, I, Chapanian, R, Higgins, M.A, Samain, E, Boraston, A.B, Kizhakkedathu, J.N, Withers, S.G. | | Deposit date: | 2014-11-19 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Towards Efficient Enzymes for the Generation of Universal Blood Through Structure-Guided Directed Evolution.
J.Am.Chem.Soc., 137, 2015
|
|
4D6J
 
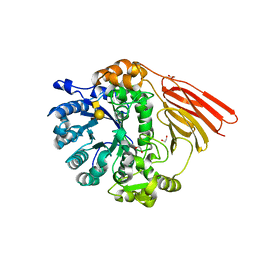 | | Crystal structure of a family 98 glycoside hydrolase catalytic module (Sp3GH98) in complex with the type 2 blood group A-tetrasaccharide (E558A X01 mutant) | | Descriptor: | 1,2-ETHANEDIOL, GLYCOSIDE HYDROLASE, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kwan, D.H, Constantinescu, I, Chapanian, R, Higgins, M.A, Samain, E, Boraston, A.B, Kizhakkedathu, J.N, Withers, S.G. | | Deposit date: | 2014-11-11 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Towards Efficient Enzymes for the Generation of Universal Blood Through Structure-Guided Directed Evolution.
J.Am.Chem.Soc., 137, 2015
|
|
4D6E
 
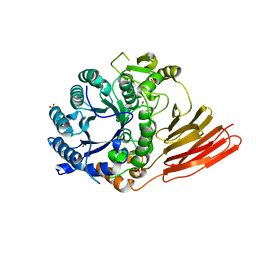 | | Crystal structure of a family 98 glycoside hydrolase catalytic module (Sp3GH98) in complex with the blood group A-trisaccharide (X01 mutant) | | Descriptor: | 1,2-ETHANEDIOL, GLYCOSIDE HYDROLASE, SULFATE ION, ... | | Authors: | Kwan, D.H, Constantinescu, I, Chapanian, R, Higgins, M.A, Samain, E, Boraston, A.B, Kizhakkedathu, J.N, Withers, S.G. | | Deposit date: | 2014-11-11 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Towards Efficient Enzymes for the Generation of Universal Blood Through Structure-Guided Directed Evolution.
J.Am.Chem.Soc., 137, 2015
|
|
4D6F
 
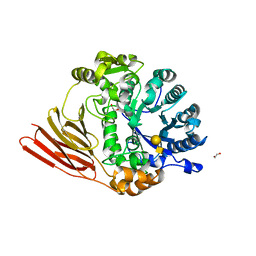 | | Crystal structure of a family 98 glycoside hydrolase catalytic module (Sp3GH98) in complex with the type 1 blood group A-tetrasaccharide (E558A, X01 mutant) | | Descriptor: | 1,2-ETHANEDIOL, GLYCOSIDE HYDROLASE, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kwan, D.H, Constantinescu, I, Chapanian, R, Higgins, M.A, Samain, E, Boraston, A.B, Kizhakkedathu, J.N, Withers, S.G. | | Deposit date: | 2014-11-11 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Towards Efficient Enzymes for the Generation of Universal Blood Through Structure-Guided Directed Evolution.
J.Am.Chem.Soc., 137, 2015
|
|
4D72
 
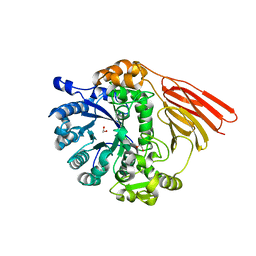 | | Crystal structure of a family 98 glycoside hydrolase catalytic module (Sp3GH98) in complex with the type 2 blood group A-tetrasaccharide (E558A L19 mutant) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, GLYCOSIDE HYDROLASE, ... | | Authors: | Kwan, D.H, Constantinescu, I, Chapanian, R, Higgins, M.A, Samain, E, Boraston, A.B, Kizhakkedathu, J.N, Withers, S.G. | | Deposit date: | 2014-11-19 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Towards Efficient Enzymes for the Generation of Universal Blood Through Structure-Guided Directed Evolution.
J.Am.Chem.Soc., 137, 2015
|
|
4D6H
 
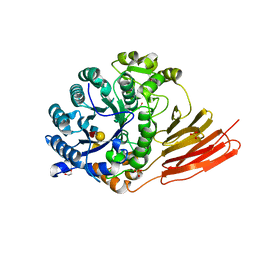 | | Crystal structure of a family 98 glycoside hydrolase catalytic module (Sp3GH98) in complex with the type 1 blood group A-tetrasaccharide (E558A X02 mutant) | | Descriptor: | 1,2-ETHANEDIOL, GLYCOSIDE HYDROLASE, SULFATE ION, ... | | Authors: | Kwan, D.H, Constantinescu, I, Chapanian, R, Higgins, M.A, Samain, E, Boraston, A.B, Kizhakkedathu, J.N, Withers, S.G. | | Deposit date: | 2014-11-11 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Towards Efficient Enzymes for the Generation of Universal Blood Through Structure-Guided Directed Evolution.
J.Am.Chem.Soc., 137, 2015
|
|
4D6I
 
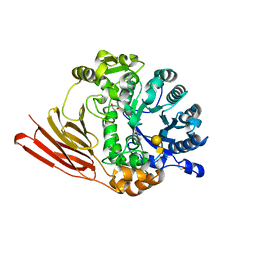 | | Crystal structure of a family 98 glycoside hydrolase catalytic module (Sp3GH98) in complex with the type 1 blood group A-tetrasaccharide (E558A L19 mutant) | | Descriptor: | 1,2-ETHANEDIOL, GLYCOSIDE HYDROLASE, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kwan, D.H, Constantinescu, I, Chapanian, R, Higgins, M.A, Samain, E, Boraston, A.B, Kizhakkedathu, J.N, Withers, S.G. | | Deposit date: | 2014-11-11 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Towards Efficient Enzymes for the Generation of Universal Blood Through Structure-Guided Directed Evolution.
J.Am.Chem.Soc., 137, 2015
|
|
4D6C
 
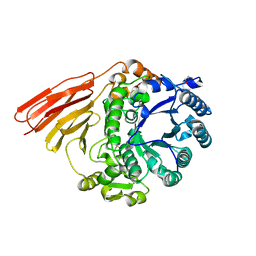 | | Crystal structure of a family 98 glycoside hydrolase catalytic module (Sp3GH98)(L19 mutant) | | Descriptor: | 1,2-ETHANEDIOL, GLYCOSIDE HYDROLASE | | Authors: | Kwan, D.H, Constantinescu, I, Chapanian, R, Higgins, M.A, Samain, E, Boraston, A.B, Kizhakkedathu, J.N, Withers, S.G. | | Deposit date: | 2014-11-11 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Towards Efficient Enzymes for the Generation of Universal Blood Through Structure-Guided Directed Evolution.
J.Am.Chem.Soc., 137, 2015
|
|
4XM5
 
 | | C. glabrata Slx1. | | Descriptor: | CHLORIDE ION, Structure-specific endonuclease subunit SLX1, ZINC ION | | Authors: | Gaur, V, Wyatt, H.D.M, Komorowska, W, Szczepanowski, R.H, de Sanctis, D, Gorecka, K.M, West, S.C, Nowotny, M. | | Deposit date: | 2015-01-14 | | Release date: | 2015-03-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural and Mechanistic Analysis of the Slx1-Slx4 Endonuclease.
Cell Rep, 10, 2015
|
|
4CC1
 
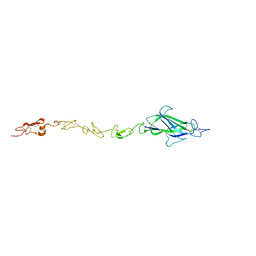 | | Notch ligand, Jagged-1, contains an N-terminal C2 domain | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Chilakuri, C.R, Sheppard, D, Ilagan, M.X.G, Holt, L.R, Abbott, F, Liang, S, Kopan, R, Handford, P.A, Lea, S.M. | | Deposit date: | 2013-10-17 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structural Analysis Uncovers Lipid-Binding Properties of Notch Ligands
Cell Rep., 5, 2013
|
|
4CBZ
 
 | | Notch ligand, Jagged-1, contains an N-terminal C2 domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN JAGGED-1, alpha-L-fucopyranose | | Authors: | Chilakuri, C.R, Sheppard, D, Ilagan, M.X.G, Holt, L.R, Abbott, F, Liang, S, Kopan, R, Handford, P.A, Lea, S.M. | | Deposit date: | 2013-10-17 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Analysis Uncovers Lipid-Binding Properties of Notch Ligands
Cell Rep., 5, 2013
|
|
4CC0
 
 | | Notch ligand, Jagged-1, contains an N-terminal C2 domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, PROTEIN JAGGED-1, ... | | Authors: | Chilakuri, C.R, Sheppard, D, Ilagan, M.X.G, Holt, L.R, Abbott, F, Liang, S, Kopan, R, Handford, P.A, Lea, S.M. | | Deposit date: | 2013-10-17 | | Release date: | 2013-11-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural Analysis Uncovers Lipid-Binding Properties of Notch Ligands
Cell Rep., 5, 2013
|
|
5AF0
 
 | | MAEL domain from Bombyx mori Maelstrom | | Descriptor: | MAELSTROM, ZINC ION | | Authors: | Chen, K, Campbell, E, Pandey, R.R, Yang, Z, McCarthy, A.A, Pillai, R.S. | | Deposit date: | 2015-01-13 | | Release date: | 2015-04-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Metazoan Maelstrom is an RNA-Binding Protein that Has Evolved from an Ancient Nuclease Active in Protists.
RNA, 21, 2015
|
|
4YEX
 
 | | HUaa-19bp | | Descriptor: | DNA-binding protein HU-alpha, synthetic DNA strand | | Authors: | Hammel, M, Reyes, F.E, Parpana, R, Tainer, J.A, Adhya, S, Amlanjyoti, D. | | Deposit date: | 2015-02-24 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | HU multimerization shift controls nucleoid compaction.
Sci Adv, 2, 2016
|
|
4YFT
 
 | | HUab-20bp | | Descriptor: | DNA-binding protein HU-alpha, DNA-binding protein HU-beta, synthetic DNA strand | | Authors: | Hammel, M, Reyes, F.E, Parpana, R, Tainer, J.A, Adhya, S, Amlanjyoti, D. | | Deposit date: | 2015-02-25 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.914 Å) | | Cite: | HU multimerization shift controls nucleoid compaction.
Sci Adv, 2, 2016
|
|
4YF0
 
 | | HU38-19bp | | Descriptor: | DNA-binding protein HU-alpha, synthetic DNA strand | | Authors: | Hammel, M, Reyes, F.E, Parpana, R, Tainer, J.A, Adhya, S, Amlanjyoti, D. | | Deposit date: | 2015-02-24 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | HU multimerization shift controls nucleoid compaction.
Sci Adv, 2, 2016
|
|
4YEW
 
 | | HUab-19bp | | Descriptor: | DNA-binding protein HU-alpha, DNA-binding protein HU-beta, synthetic DNA strand | | Authors: | Hammel, M, Reyes, F.E, Parpana, R, Tainer, J.A, Adhya, S, Amlanjyoti, D. | | Deposit date: | 2015-02-24 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.683 Å) | | Cite: | HU multimerization shift controls nucleoid compaction.
Sci Adv, 2, 2016
|
|
4YEY
 
 | | HUaa-20bp | | Descriptor: | DNA-binding protein HU-alpha, synthetic DNA strand | | Authors: | Hammel, M, Reyes, F.E, Parpana, R, Tainer, J.A, Adhya, S, Amlanjyoti, D. | | Deposit date: | 2015-02-24 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.354 Å) | | Cite: | HU multimerization shift controls nucleoid compaction.
Sci Adv, 2, 2016
|
|
