1QNU
 
 | | Shiga-Like Toxin I B Subunit Complexed with the Bridged-Starfish Inhibitor | | Descriptor: | ETHYL-CARBAMIC ACID METHYL ESTER, METHYL-CARBAMIC ACID ETHYL ESTER, Shiga toxin 1 variant B subunit, ... | | Authors: | Pannu, N.S, Hayakawa, K, Read, R.J. | | Deposit date: | 1999-10-21 | | Release date: | 2000-04-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Shiga-like toxins are neutralized by tailored multivalent carbohydrate ligands.
Nature, 403, 2000
|
|
1QOH
 
 | | A MUTANT SHIGA-LIKE TOXIN IIE | | Descriptor: | SHIGA-LIKE TOXIN IIE B SUBUNIT | | Authors: | Pannu, N.S, Boodhoo, A, Armstrong, G.D, Clark, C.G, Brunton, J.L, Read, R.J. | | Deposit date: | 1999-11-08 | | Release date: | 2000-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A Mutant Shiga-Like Toxin Iie Bound to its Receptor Gb(3): Structure of a Group II Shiga-Like Toxin with Altered Binding Specificity
Structure, 8, 2000
|
|
7CCA
 
 | | Crystal structure of White Spot Syndrome Virus Thymidylate Synthase - ternary complex with Methotrexate and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, METHOTREXATE, Thymidylate Synthase | | Authors: | Panchal, N.V, Kumar, S, Shaikh, N, Vasudevan, D. | | Deposit date: | 2020-06-16 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure analysis of thymidylate synthase from white spot syndrome virus reveals WSSV-specific structural elements.
Int.J.Biol.Macromol., 167, 2021
|
|
7WJL
 
 | | Crystal structure of S. cerevisiae Hos3 | | Descriptor: | ACETATE ION, Histone deacetylase HOS3, ZINC ION | | Authors: | Pang, N.N, Che, S.Y, Yang, N. | | Deposit date: | 2022-01-07 | | Release date: | 2023-01-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural characterization of fungus-specific histone deacetylase Hos3 provides insights into developing selective inhibitors with antifungal activity.
J.Biol.Chem., 298, 2022
|
|
5DU3
 
 | | Active form of human C1-inhibitor | | Descriptor: | Plasma protease C1 inhibitor | | Authors: | Pannu, N.S, Dijk, M, Holkers, J, Voskamp, P, Giannetti, B.M, Waterreus, W.J, van Veen, H.A. | | Deposit date: | 2015-09-18 | | Release date: | 2016-08-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | How Dextran Sulfate Affects C1-inhibitor Activity: A Model for Polysaccharide Potentiation.
Structure, 24, 2016
|
|
4H0K
 
 | | Mutant m58h of Nostoc sp cytochrome c6 | | Descriptor: | Cytochrome c6, HEME C | | Authors: | Pannu, N.S, Skubak, P, Cavazzini, D, Rossi, G.L, Ubbink, M. | | Deposit date: | 2012-09-08 | | Release date: | 2013-09-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The dynamic complex of cytochrome c6 and cytochrome f studied with paramagnetic NMR spectroscopy
Biochim.Biophys.Acta, 1837, 2014
|
|
4H0J
 
 | | Mutant M58C of Nostoc sp Cytochrome c6 | | Descriptor: | Cytochrome c6, HEME C | | Authors: | Pannu, N.S, Skubak, P, Ubbink, M, Cavazzini, D, Rossi, G.L. | | Deposit date: | 2012-09-08 | | Release date: | 2013-09-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The dynamic complex of cytochrome c6 and cytochrome f studied with paramagnetic NMR spectroscopy
Biochim.Biophys.Acta, 1837, 2014
|
|
4GM7
 
 | | Structure of cinnamic acid bound bovine lactoperoxidase at 2.6A resolution. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pandey, N, Singh, A.K, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-08-15 | | Release date: | 2012-09-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of cinnamic acid bound bovine lactoperoxidase at 2.6A resolution
To be Published
|
|
2LMA
 
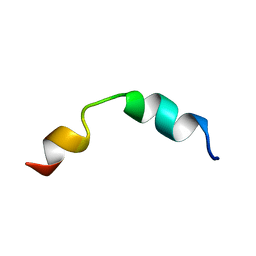 | | Solution structure of CD4+ T cell derived peptide Thp5 | | Descriptor: | Thp5 peptide | | Authors: | Pandey, N.K, Khan, M.M, Chatterjee, S, Dwivedi, V.P, Tousif, S, Das, J, Van Kaer, L. | | Deposit date: | 2011-11-29 | | Release date: | 2011-12-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | CD4+ T cell derived novel peptide Thp5 induces IL-4 production in CD4+ T cells to direct T helper 2 cell differentiation
J.Biol.Chem., 2011
|
|
5V3S
 
 | |
5ZDL
 
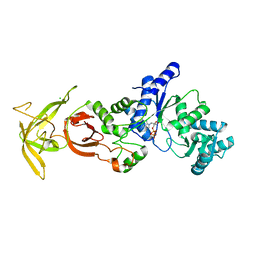 | | Crystal Structure Analysis of TtQRS in co-crystallised with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, Glutamine--tRNA ligase | | Authors: | Mutharasappan, N, Jain, V, Sharma, A, Manickam, Y, Jeyaraman, J. | | Deposit date: | 2018-02-23 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional analysis of Glutaminyl-tRNA synthetase (TtGlnRS) from Thermus thermophilus HB8 and its complexes
Int. J. Biol. Macromol., 120, 2018
|
|
5ZDO
 
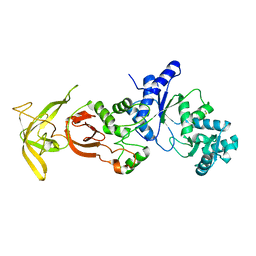 | | Crystal Structure Analysis of TtQRS in co-crystallised with ATP | | Descriptor: | CHLORIDE ION, Glutamine-tRNA ligase | | Authors: | Mutharasappan, N, Jain, V, Sharma, A, Manickam, Y, Jeyaraman, J. | | Deposit date: | 2018-02-23 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and functional analysis of Glutaminyl-tRNA synthetase (TtGlnRS) from Thermus thermophilus HB8 and its complexes
Int. J. Biol. Macromol., 120, 2018
|
|
5ZG8
 
 | |
3JTI
 
 | | Crystal structure of the complex formed between Phospholipase A2 with beta-amyloid fragment, Lys-Gly-Ala-Ile-Ile-Gly-Leu-Met at 1.8 A resolution | | Descriptor: | CALCIUM ION, Phospholipase A2 isoform 3, octapeptide from Amyloid beta A4 protein | | Authors: | Pandey, N, Mirza, Z, Vikram, G, Singh, N, Bhushan, A, Kaur, P, Srinivasan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2009-09-12 | | Release date: | 2010-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the complex formed between Phospholipase A2 with beta-amyloid fragment, Lys-Gly-Ala-Ile-Ile-Gly-Leu-Met at 1.8 A resolution
To be Published
|
|
5ZDK
 
 | | Crystal Structure Analysis of TtQRS in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CESIUM ION, CHLORIDE ION, ... | | Authors: | Mutharasappan, N, Jain, V, Sharma, A, Manickam, Y, Jeyaraman, J. | | Deposit date: | 2018-02-23 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural and functional analysis of Glutaminyl-tRNA synthetase (TtGlnRS) from Thermus thermophilus HB8 and its complexes
Int. J. Biol. Macromol., 120, 2018
|
|
5IHF
 
 | |
5GHU
 
 | | Crystal structure of YadV chaperone at 1.63 Angstrom | | Descriptor: | ACETATE ION, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Pandey, N.K, Bhavesh, N.S. | | Deposit date: | 2016-06-20 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structure of the usher chaperone YadV reveals a monomer with the proline lock in closed conformation suggestive of an intermediate state.
Febs Lett., 2020
|
|
5IO8
 
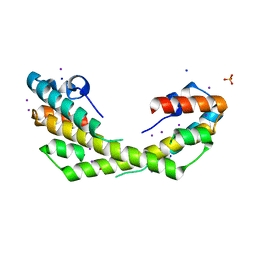 | | Salmonella Typhimurium VirG-like (STV) protein at 2.19 Angstrom resolution solved by Iodine SAD. | | Descriptor: | IODIDE ION, PHOSPHATE ION, SODIUM ION, ... | | Authors: | Pandey, N.K, Ray, S, Suar, M, Bhavesh, N.S. | | Deposit date: | 2016-03-08 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.192 Å) | | Cite: | Salmonella Typhimurium VirG-like (STV) protein at at 2.19 Angstrom resolution by SAD.
To Be Published
|
|
3N1D
 
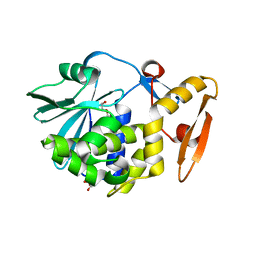 | | Crystal structure of the complex of type I ribosome inactivating protein with ribose at 1.7A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Pandey, N, Kushwaha, G.S, Sinha, M, Kaur, P, Betzel, C, Sharma, S, Singh, T.P. | | Deposit date: | 2010-05-15 | | Release date: | 2010-06-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the complex of type I ribosome inactivating protein with ribose at 1.7A resolution
To be Published
|
|
3N5D
 
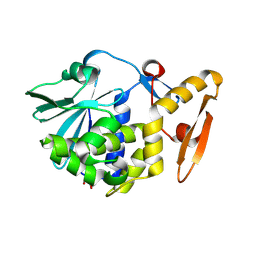 | | Crystal structure of the complex of type I ribosome inactivating protein with glucose at 1.9A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Ribosome inactivating protein, ... | | Authors: | Pandey, N, Kushwaha, G.S, Sinha, M, Kaur, P, Betzel, C, Sharma, S, Singh, T.P. | | Deposit date: | 2010-05-25 | | Release date: | 2010-06-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the complex of type I ribosome inactivating protein with glucose at 1.9A resolution
To be Published
|
|
3NJS
 
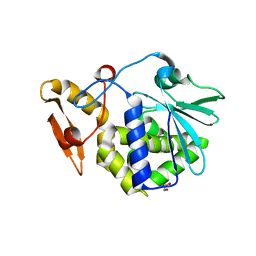 | | Crystal structure of the complex formed between typeI ribosome inactivating protein and lactose at 2.1A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Ribosome inactivating protein, ... | | Authors: | Pandey, N, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-06-18 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the complex formed between typeI ribosome inactivating protein and lactose at 2.1A resolution
To be Published
|
|
3UMQ
 
 | | Crystal structure of peptidoglycan recognition protein-S complexed with butyric acid at 2.2 A resolution | | Descriptor: | GLYCEROL, Peptidoglycan recognition protein 1, butanoic acid | | Authors: | Pandey, N, Sharma, P, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-11-14 | | Release date: | 2012-07-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of the binding of fatty acids to peptidoglycan recognition protein, PGRP-S through second binding site
Arch.Biochem.Biophys., 529, 2013
|
|
3U59
 
 | |
3NX9
 
 | | Crystal structure of type I ribosome inactivating protein in complex with maltose at 1.7A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Ribosome inactivating protein, ... | | Authors: | Pandey, N, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-07-13 | | Release date: | 2010-08-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of type I ribosome inactivating protein in complex with maltose at 1.7A resolution
To be Published
|
|
3U1A
 
 | |
