8I67
 
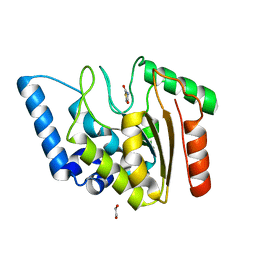 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with 2,4-Thiazolidinedione, Form I | | Descriptor: | 1,2-ETHANEDIOL, 1,3-thiazolidine-2,4-dione, Uracil-DNA glycosylase | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment.
Eur.J.Med.Chem., 258, 2023
|
|
8I69
 
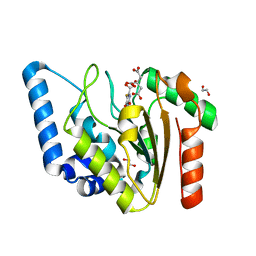 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with 5-Fluoroorotic acid and Citric acid, Form I | | Descriptor: | 1,2-ETHANEDIOL, 5-FLUORO-2,6-DIOXO-1,2,3,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, CITRIC ACID, ... | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment.
Eur.J.Med.Chem., 258, 2023
|
|
8I6B
 
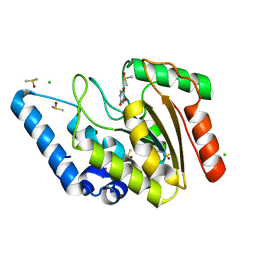 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with 5-Hydroxy-2,4(1H,3H)-pyrimidinedione, Form I | | Descriptor: | 1,2-ETHANEDIOL, 5-oxidanyl-1~{H}-pyrimidine-2,4-dione, CHLORIDE ION, ... | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment.
Eur.J.Med.Chem., 258, 2023
|
|
8I63
 
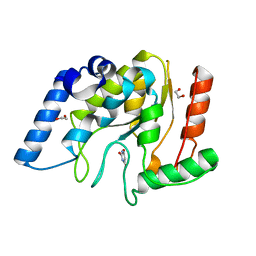 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with Barbituric acid, Form III | | Descriptor: | 1,2-ETHANEDIOL, BARBITURIC ACID, Uracil-DNA glycosylase | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment.
Eur.J.Med.Chem., 258, 2023
|
|
8I66
 
 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with isoorotic acid (2,4-Dihydroxypyrimidine-5-carboxylic Acid) and citric acid, Form I | | Descriptor: | 2,4-dioxo-1,2,3,4-tetrahydropyrimidine-5-carboxylic acid, CITRIC ACID, Uracil-DNA glycosylase | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment.
Eur.J.Med.Chem., 258, 2023
|
|
8I62
 
 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with Barbituric acid, Form I | | Descriptor: | 1,2-ETHANEDIOL, BARBITURIC ACID, CHLORIDE ION, ... | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment.
Eur.J.Med.Chem., 258, 2023
|
|
8I65
 
 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with isoorotic acid (2,4-Dihydroxypyrimidine-5-carboxylic Acid), Form I | | Descriptor: | 1,2-ETHANEDIOL, 2,4-dioxo-1,2,3,4-tetrahydropyrimidine-5-carboxylic acid, Uracil-DNA glycosylase | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment.
Eur.J.Med.Chem., 258, 2023
|
|
8I6A
 
 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with Orotic acid, Form III | | Descriptor: | 1,2-ETHANEDIOL, OROTIC ACID, Uracil-DNA glycosylase | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment.
Eur.J.Med.Chem., 258, 2023
|
|
8I6C
 
 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with 6-Formyl-uracil, Form III | | Descriptor: | 6-[bis(oxidanyl)methyl]-5~{H}-pyrimidine-2,4-dione, Uracil-DNA glycosylase | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment.
Eur.J.Med.Chem., 258, 2023
|
|
8I6D
 
 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with 5-Hydroxy-2,4(1H,3H)-pyrimidinedione, Form VI | | Descriptor: | 1,2-ETHANEDIOL, 5-oxidanyl-1~{H}-pyrimidine-2,4-dione, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment.
Eur.J.Med.Chem., 258, 2023
|
|
8I64
 
 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with Barbituric acid, Form II | | Descriptor: | 1,2-ETHANEDIOL, BARBITURIC ACID, Uracil-DNA glycosylase | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment.
Eur.J.Med.Chem., 258, 2023
|
|
8I68
 
 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with Uric acid, Form III | | Descriptor: | 1,2-ETHANEDIOL, URIC ACID, Uracil-DNA glycosylase | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment.
Eur.J.Med.Chem., 258, 2023
|
|
5ITW
 
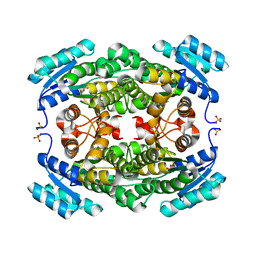 | | Crystal structure of Bacillus subtilis BacC Dihydroanticapsin 7-dehydrogenase | | Descriptor: | Dihydroanticapsin 7-dehydrogenase, SULFATE ION | | Authors: | Perinbam, K, Balaram, H, Row, T.N.G, Gopal, B. | | Deposit date: | 2016-03-17 | | Release date: | 2017-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Probing the influence of non-covalent contact networks identified by charge density analysis on the oxidoreductase BacC.
Protein Eng. Des. Sel., 30, 2017
|
|
5ITV
 
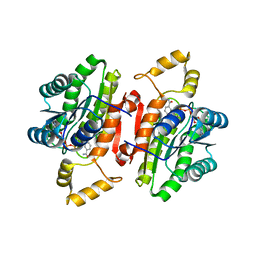 | | Crystal structure of Bacillus subtilis BacC Dihydroanticapsin 7-dehydrogenase in complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Dihydroanticapsin 7-dehydrogenase | | Authors: | Perinbam, K, Balaram, H, Row, T.N.G, Gopal, B. | | Deposit date: | 2016-03-17 | | Release date: | 2017-02-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Probing the influence of non-covalent contact networks identified by charge density analysis on the oxidoreductase BacC.
Protein Eng. Des. Sel., 30, 2017
|
|
7ESS
 
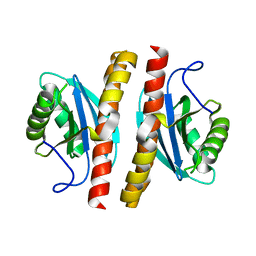 | | Structure-guided studies of the Holliday junction resolvase RuvX provide novel insights into ATP-stimulated cleavage of branched DNA and RNA substrates | | Descriptor: | Putative pre-16S rRNA nuclease | | Authors: | Thakur, M, Mohan, D, Singh, A.K, Agarwal, A, Gopal, B, Muniyappa, K. | | Deposit date: | 2021-05-11 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Novel insights into ATP-Stimulated Cleavage of branched DNA and RNA Substrates through Structure-Guided Studies of the Holliday Junction Resolvase RuvX.
J.Mol.Biol., 433, 2021
|
|
7F5Y
 
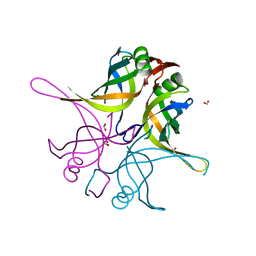 | | Crystal structure of the single-stranded dna-binding protein from Mycobacterium tuberculosis- Form III | | Descriptor: | FORMIC ACID, Single-stranded DNA-binding protein | | Authors: | Srikalaivani, R, Paul, A, Sriram, R, Narayanan, S, Gopal, B, Vijayan, M. | | Deposit date: | 2021-06-23 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural variability of Mycobacterium tuberculosis SSB and susceptibility to inhibition.
Curr.Sci., 122, 2022
|
|
7F5Z
 
 | | Crystal structure of the single-stranded dna-binding protein from Mycobacterium tuberculosis- Form III | | Descriptor: | Single-stranded DNA-binding protein | | Authors: | Srikalaivani, R, Paul, A, Sriram, R, Narayanan, S, Gopal, B, Vijayan, M. | | Deposit date: | 2021-06-23 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural variability of Mycobacterium tuberculosis SSB and susceptibility to inhibition.
Curr.Sci., 122, 2022
|
|
7W22
 
 | | Structure of the M. tuberculosis HtrA K436A mutant | | Descriptor: | Probable serine protease HtrA1 | | Authors: | Gupta, A.K, Gopal, B. | | Deposit date: | 2021-11-21 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Allosteric Determinants in High Temperature Requirement A Enzymes Are Conserved and Regulate the Population of Active Conformations.
Acs Chem.Biol., 18, 2023
|
|
7W23
 
 | | Structure of the M. tuberculosis HtrA S363A mutant | | Descriptor: | Probable serine protease HtrA1 | | Authors: | Gupta, A.K, Gopal, B. | | Deposit date: | 2021-11-21 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Allosteric Determinants in High Temperature Requirement A Enzymes Are Conserved and Regulate the Population of Active Conformations.
Acs Chem.Biol., 18, 2023
|
|
7W24
 
 | | Structure of the M. tuberculosis HtrA N383A mutant | | Descriptor: | Probable serine protease HtrA1 | | Authors: | Gupta, A.K, Gopal, B. | | Deposit date: | 2021-11-21 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Allosteric Determinants in High Temperature Requirement A Enzymes Are Conserved and Regulate the Population of Active Conformations.
Acs Chem.Biol., 18, 2023
|
|
7W25
 
 | | Structure of the M. tuberculosis HtrA S413A mutant | | Descriptor: | Probable serine protease HtrA1 | | Authors: | Gupta, A.K, Gopal, B. | | Deposit date: | 2021-11-21 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Allosteric Determinants in High Temperature Requirement A Enzymes Are Conserved and Regulate the Population of Active Conformations.
Acs Chem.Biol., 18, 2023
|
|
7VZ0
 
 | | Structure of the M. tuberculosis HtrA S407A mutant | | Descriptor: | Probable serine protease HtrA1 | | Authors: | Gupta, A.K, Gopal, B. | | Deposit date: | 2021-11-15 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Allosteric Determinants in High Temperature Requirement A Enzymes Are Conserved and Regulate the Population of Active Conformations.
Acs Chem.Biol., 18, 2023
|
|
7W21
 
 | | Structure of the M. tuberculosis HtrA N269A mutant | | Descriptor: | Probable serine protease HtrA1 | | Authors: | Gupta, A.K, Gopal, B. | | Deposit date: | 2021-11-21 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Allosteric Determinants in High Temperature Requirement A Enzymes Are Conserved and Regulate the Population of Active Conformations.
Acs Chem.Biol., 18, 2023
|
|
7VYZ
 
 | | Structure of the M. tuberculosis HtrA S367A mutant | | Descriptor: | Probable serine protease HtrA1 | | Authors: | Gupta, A.K, Gopal, B. | | Deposit date: | 2021-11-15 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Allosteric Determinants in High Temperature Requirement A Enzymes Are Conserved and Regulate the Population of Active Conformations.
Acs Chem.Biol., 18, 2023
|
|
7W4R
 
 | |
