6OJY
 
 | |
7ZUJ
 
 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) in complex with lactone 6Az - Streptococcus pneumoniae R6 | | Descriptor: | 6-azido-N-[(2S)-1-oxidanylidene-1-[[(2S,3R)-3-oxidanyl-1-oxidanylidene-butan-2-yl]amino]propan-2-yl]hexanamide, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Flanders, P.L, Contreras-Martel, C, Martins, A, Brown, N.W, Shirley, J.D, Nauta, K.M, Dessen, A, Carlson, E.E, Ambrose, E.A. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Combined Structural Analysis and Molecular Dynamics Reveal Penicillin-Binding Protein Inhibition Mode with beta-Lactones.
Acs Chem.Biol., 17, 2022
|
|
7ZUK
 
 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) in complex with lactone 7Az - Streptococcus pneumoniae R6 | | Descriptor: | 6-azido-N-[(2S)-1-oxidanylidene-1-[[(2S,3R)-3-oxidanyl-1-oxidanylidene-butan-2-yl]amino]-3-phenyl-propan-2-yl]hexanamide, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Flanders, P.L, Contreras-Martel, C, Martins, A, Brown, N.W, Shirley, J.D, Nauta, K.M, Dessen, A, Carlson, E.E, Ambrose, E.A. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | Combined Structural Analysis and Molecular Dynamics Reveal Penicillin-Binding Protein Inhibition Mode with beta-Lactones.
Acs Chem.Biol., 17, 2022
|
|
7ZUL
 
 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) in complex with 8Az lactone - Streptococcus pneumoniae R6 | | Descriptor: | 6-azido-N-[(2R)-1-oxidanylidene-1-[[(2S,3R)-3-oxidanyl-1-oxidanylidene-butan-2-yl]amino]-3-phenyl-propan-2-yl]hexanamide, CHLORIDE ION, Penicillin-binding protein 1b | | Authors: | Flanders, P.L, Contreras-Martel, C, Martins, A, Brown, N.W, Shirley, J.D, Nauta, K.M, Dessen, A, Carlson, E.E, Ambrose, E.A. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.744 Å) | | Cite: | Combined Structural Analysis and Molecular Dynamics Reveal Penicillin-Binding Protein Inhibition Mode with beta-Lactones.
Acs Chem.Biol., 17, 2022
|
|
5E7C
 
 | | Macromolecular diffractive imaging using imperfect crystals - Bragg data | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Ayyer, K, Yefanov, O, Oberthuer, D, Roy-Chowdhury, S, Galli, L, Mariani, V, Basu, S, Coe, J, Conrad, C.E, Fromme, R, Schaffner, A, Doerner, K, James, D, Kupitz, C, Metz, M, Nelson, G, Xavier, P.L, Beyerlein, K.R, Schmidt, M, Sarrou, I, Spence, J.C.H, Weierstall, U, White, T.A, Yang, J.-H, Zhao, Y, Liang, M, Aquila, A, Hunter, M.S, Robinson, J.S, Koglin, J.E, Boutet, S, Fromme, P, Barty, A, Chapman, H.N. | | Deposit date: | 2015-10-12 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Macromolecular diffractive imaging using imperfect crystals.
Nature, 530, 2016
|
|
6OJ1
 
 | | Crystal Structure of Aspergillus fumigatus Ega3 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bamford, N.C, Subramanian, A.S, Millan, C, Uson, I, Howell, P.L. | | Deposit date: | 2019-04-10 | | Release date: | 2019-08-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Ega3 from the fungal pathogenAspergillus fumigatusis an endo-alpha-1,4-galactosaminidase that disrupts microbial biofilms.
J.Biol.Chem., 294, 2019
|
|
6CHG
 
 | | Crystal structure of the yeast COMPASS catalytic module | | Descriptor: | H3, Histone-lysine N-methyltransferase, H3 lysine-4 specific, ... | | Authors: | Hsu, P.L, Li, H, Zheng, N. | | Deposit date: | 2018-02-22 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.985 Å) | | Cite: | Crystal Structure of the COMPASS H3K4 Methyltransferase Catalytic Module.
Cell, 174, 2018
|
|
6NWZ
 
 | | Crystal structure of Agd3 a novel carbohydrate deacetylase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Bamford, N.C, Howell, P.L. | | Deposit date: | 2019-02-07 | | Release date: | 2020-02-12 | | Last modified: | 2020-08-26 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and biochemical characterization of the exopolysaccharide deacetylase Agd3 required for Aspergillus fumigatus biofilm formation.
Nat Commun, 11, 2020
|
|
7ZUI
 
 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) in complex with lactone 5Az - Streptococcus pneumoniae R6 | | Descriptor: | 6-azido-N-[(2R)-1-oxidanylidene-1-[[(2S,3R)-3-oxidanyl-1-oxidanylidene-butan-2-yl]amino]propan-2-yl]hexanamide, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Flanders, P.L, Contreras-Martel, C, Martins, A, Brown, N.W, Shirley, J.D, Nauta, K.M, Dessen, A, Carlson, E.E, Ambrose, E.A. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Combined Structural Analysis and Molecular Dynamics Reveal Penicillin-Binding Protein Inhibition Mode with beta-Lactones.
Acs Chem.Biol., 17, 2022
|
|
7ZUH
 
 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) Streptococcus pneumoniae R6 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Penicillin-binding protein 1b | | Authors: | Flanders, P.L, Contreras-Martel, C, Martins, A, Brown, N.W, Shirley, J.D, Nauta, K.M, Dessen, A, Carlson, E.E, Ambrose, E.A. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.467 Å) | | Cite: | Combined Structural Analysis and Molecular Dynamics Reveal Penicillin-Binding Protein Inhibition Mode with beta-Lactones.
Acs Chem.Biol., 17, 2022
|
|
6OJX
 
 | |
6OKV
 
 | |
5EQ6
 
 | | Pseudomonas aeruginosa PilM bound to AMP-PNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Type 4 fimbrial biogenesis protein PilM | | Authors: | McCallum, M, Tammam, S, Robinson, H, Shah, M, Calmettes, C, Moraes, T, Burrows, L.L, Howell, P.L. | | Deposit date: | 2015-11-12 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | PilN Binding Modulates the Structure and Binding Partners of the Pseudomonas aeruginosa Type IVa Pilus Protein PilM.
J.Biol.Chem., 291, 2016
|
|
6MSI
 
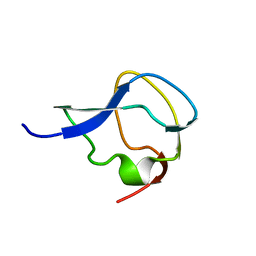 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 | | Descriptor: | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 | | Authors: | Deluca, C.I, Davies, P.L, Ye, Q, Jia, Z. | | Deposit date: | 1997-09-17 | | Release date: | 1998-10-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The effects of steric mutations on the structure of type III antifreeze protein and its interaction with ice.
J.Mol.Biol., 275, 1998
|
|
6D10
 
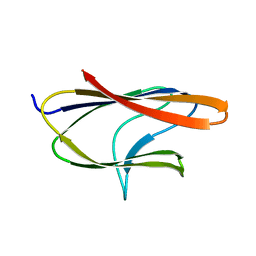 | |
5EOY
 
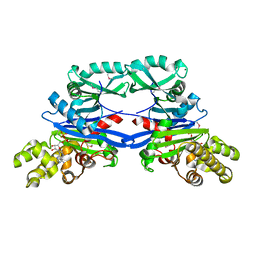 | | Pseudomonas aeruginosa SeMet-PilM bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Type 4 fimbrial biogenesis protein PilM | | Authors: | McCallum, M, Tammam, S, Robinson, H, Shah, M, Calmettes, C, Moraes, T, Burrows, L.L, Howell, P.L. | | Deposit date: | 2015-11-10 | | Release date: | 2016-04-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | PilN Binding Modulates the Structure and Binding Partners of the Pseudomonas aeruginosa Type IVa Pilus Protein PilM.
J.Biol.Chem., 291, 2016
|
|
6OLK
 
 | |
6OJB
 
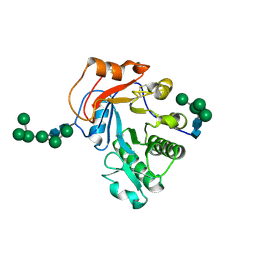 | | Crystal Structure of Aspergillus fumigatus Ega3 complex with galactosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-2-deoxy-alpha-D-galactopyranose, ... | | Authors: | Bamford, N.C, Howell, P.L. | | Deposit date: | 2019-04-11 | | Release date: | 2019-08-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Ega3 from the fungal pathogenAspergillus fumigatusis an endo-alpha-1,4-galactosaminidase that disrupts microbial biofilms.
J.Biol.Chem., 294, 2019
|
|
6OLM
 
 | |
5EOX
 
 | | Pseudomonas aeruginosa PilM bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Type 4 fimbrial biogenesis protein PilM | | Authors: | McCallum, M, Tammam, S, Robinson, H, Shah, M, Calmettes, C, Moraes, T, Burrows, L.L, Howell, P.L. | | Deposit date: | 2015-11-10 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | PilN Binding Modulates the Structure and Binding Partners of the Pseudomonas aeruginosa Type IVa Pilus Protein PilM.
J.Biol.Chem., 291, 2016
|
|
5D6T
 
 | | Crystal Structure of Aspergillus clavatus Sph3 in complex with GalNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, CHLORIDE ION, SPHERULIN-4, ... | | Authors: | Bamford, N.C, Little, D.J, Howell, P.L. | | Deposit date: | 2015-08-12 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Sph3 Is a Glycoside Hydrolase Required for the Biosynthesis of Galactosaminogalactan in Aspergillus fumigatus.
J.Biol.Chem., 290, 2015
|
|
5BXA
 
 | | Structure of PslG from Pseudomonas aeruginosa in complex with mannose | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Baker, P, Little, D.J, Howell, P.L. | | Deposit date: | 2015-06-08 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization of the Pseudomonas aeruginosa Glycoside Hydrolase PslG Reveals That Its Levels Are Critical for Psl Polysaccharide Biosynthesis and Biofilm Formation.
J.Biol.Chem., 290, 2015
|
|
2QTG
 
 | | Crystal Structure of Arabidopsis thaliana 5'-Methylthioadenosine nucleosidase in complex with 5'-methylthiotubercidin | | Descriptor: | 1,2-ETHANEDIOL, 2-(4-AMINO-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-METHYLSULFANYLMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, 5'-methylthioadenosine nucleosidase | | Authors: | Siu, K.K.W, Howell, P.L. | | Deposit date: | 2007-08-02 | | Release date: | 2008-04-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Molecular determinants of substrate specificity in plant 5'-methylthioadenosine nucleosidases.
J.Mol.Biol., 378, 2008
|
|
2QTT
 
 | | Crystal Structure of Arabidopsis thaliana 5'-Methylthioadenosine nucleosidase in complex with Formycin A | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, 1,2-ETHANEDIOL, 5'-methylthioadenosine nucleosidase, ... | | Authors: | Siu, K.K.W, Howell, P.L. | | Deposit date: | 2007-08-02 | | Release date: | 2008-04-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Molecular determinants of substrate specificity in plant 5'-methylthioadenosine nucleosidases.
J.Mol.Biol., 378, 2008
|
|
4F9D
 
 | | Structure of Escherichia coli PgaB 42-655 in complex with nickel | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETIC ACID, CALCIUM ION, ... | | Authors: | Little, D.J, Poloczek, J, Whitney, J.C, Robinson, H, Nitz, M, Howell, P.L. | | Deposit date: | 2012-05-18 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structure and Metal Dependent Activity of Escherichia coli PgaB Provides Insight into the Partial De-N-acetylation of Poly-b-1,6-N-acetyl-D-glucosamine
J.Biol.Chem., 287, 2012
|
|
