8VW4
 
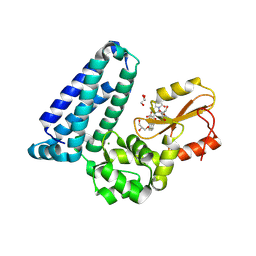 | | Crystal structure of Cbl-b TKB bound to compound 26 | | Descriptor: | (7-methoxy-2-{2-[(1S,3S,4S)-3-(3-methoxy-2-methyl-5-nitrophenyl)-1-methyl-5-oxo-1,5-dihydroimidazo[1,5-a]pyridin-2(3H)-yl]-2-oxoethoxy}quinolin-8-yl)acetic acid, DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase CBL-B, ... | | Authors: | Yu, C, Murray, J, Hsu, P.L. | | Deposit date: | 2024-01-31 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Optimization of a Novel DEL Hit That Binds in the Cbl-b SH2 Domain and Blocks Substrate Binding.
Acs Med.Chem.Lett., 15, 2024
|
|
6OK2
 
 | |
6OD6
 
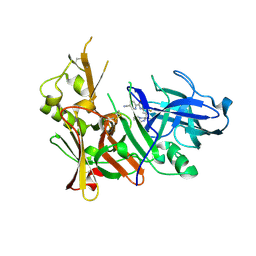 | | Structure of BACE-1 in complex with Ligand 13 | | Descriptor: | Beta-secretase 1, GLYCEROL, N-{3-[(3R)-1-amino-3-methyl-3,4-dihydropyrrolo[1,2-a]pyrazin-3-yl]-4-fluorophenyl}-5-cyanopyridine-2-carboxamide | | Authors: | Shaffer, P.L. | | Deposit date: | 2019-03-26 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evaluation of a Series of beta-Secretase 1 Inhibitors Containing Novel Heteroaryl-Fused-Piperazine Amidine Warheads.
Acs Med.Chem.Lett., 10, 2019
|
|
6OLL
 
 | |
4AME
 
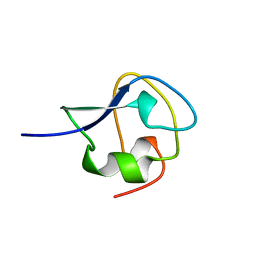 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 T18A | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
6OJZ
 
 | |
4CHV
 
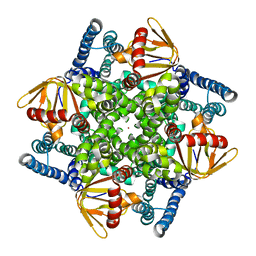 | | The electron crystallography structure of the cAMP-bound potassium channel MloK1 | | Descriptor: | CYCLIC NUCLEOTIDE-GATED POTASSIUM CHANNEL MLL3241, POTASSIUM ION | | Authors: | Kowal, J, Chami, M, Baumgartner, P, Arheit, M, Chiu, P.L, Rangl, M, Scheuring, S, Schroeder, G.F, Nimigean, C.M, Stahlberg, H. | | Deposit date: | 2013-12-04 | | Release date: | 2014-01-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON CRYSTALLOGRAPHY (7 Å) | | Cite: | Ligand-Induced Structural Changes in the Cyclic Nucleotide-Modulated Potassium Channel Mlok1
Nat.Commun., 5, 2014
|
|
9AME
 
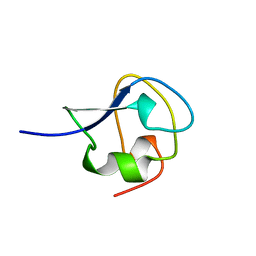 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 S42G | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
4DN0
 
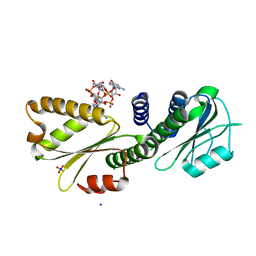 | | PelD 156-455 from Pseudomonas aeruginosa PA14 in complex with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Putative uncharacterized protein pelD, SODIUM ION, ... | | Authors: | Whitney, J.C, Colvin, K.M, Marmont, L.S, Robinson, H, Parsek, M.R, Howell, P.L. | | Deposit date: | 2012-02-08 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Cytoplasmic Region of PelD, a Degenerate Diguanylate Cyclase Receptor That Regulates Exopolysaccharide Production in Pseudomonas aeruginosa.
J.Biol.Chem., 287, 2012
|
|
4DJK
 
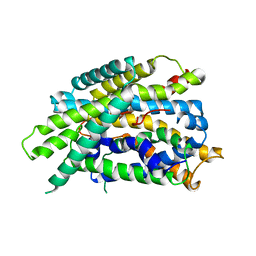 | | Structure of glutamate-GABA antiporter GadC | | Descriptor: | Probable glutamate/gamma-aminobutyrate antiporter | | Authors: | Ma, D, Lu, P.L, Yan, C.Y, Fan, C, Yin, P, Wang, J.W, Shi, Y.G. | | Deposit date: | 2012-02-02 | | Release date: | 2012-03-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.097 Å) | | Cite: | Structure and mechanism of a glutamate-GABA antiporter
Nature, 483, 2012
|
|
1QH7
 
 | | CATALYSIS AND SPECIFICITY IN ENZYMATIC GLYCOSIDE HYDROLASES: A 2,5B CONFORMATION FOR THE GLYCOSYL-ENZYME INTERMIDIATE REVEALED BY THE STRUCTURE OF THE BACILLUS AGARADHAERENS FAMILY 11 XYLANASE | | Descriptor: | XYLANASE, beta-D-xylopyranose | | Authors: | Sabini, E, Sulzenbacher, G, Dauter, M, Dauter, Z, Jorgensen, P.L, Schulein, M, Dupont, C, Davies, G.J, Wilson, K.S. | | Deposit date: | 1999-05-11 | | Release date: | 2000-05-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Catalysis and specificity in enzymatic glycoside hydrolysis: a 2,5B conformation for the glycosyl-enzyme intermediate revealed by the structure of the Bacillus agaradhaerens family 11 xylanase.
Chem.Biol., 6, 1999
|
|
5FNQ
 
 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | Descriptor: | 3-(4-CHLOROPHENYL)PROPANOIC ACID, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | Authors: | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | Deposit date: | 2015-11-16 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
5FZN
 
 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | Descriptor: | KELCH-LIKE ECH-ASSOCIATED PROTEIN 1, SULFATE ION, benzenesulfonamide | | Authors: | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | Deposit date: | 2016-03-15 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
5FNT
 
 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | Descriptor: | (3S)-3-{4-Chloro-3-[(N-methylbenzenesulfonamido) methyl]phenyl}-3-(1-methyl-1H-1,2,3-benzotriazol-5-yl)propanoic acid, CHLORIDE ION, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | Authors: | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | Deposit date: | 2015-11-16 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
5FZJ
 
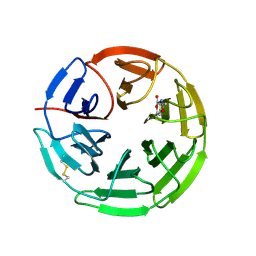 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | Descriptor: | 2,6-DIMETHYL-4H-PYRANO[3,4-D][1,3]OXAZOL-4-ONE, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | Authors: | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | Deposit date: | 2016-03-14 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
1QFT
 
 | | HISTAMINE BINDING PROTEIN FROM FEMALE BROWN EAR RHIPICEPHALUS APPENDICULATUS | | Descriptor: | HISTAMINE, PROTEIN (FEMALE-SPECIFIC HISTAMINE BINDING PROTEIN 2) | | Authors: | Paesen, G.C, Adams, P.L, Harlos, K, Nuttal, P.A, Stuart, D.I. | | Deposit date: | 1999-04-14 | | Release date: | 2000-04-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Tick histamine-binding proteins: isolation, cloning, and three-dimensional structure.
Mol.Cell, 3, 1999
|
|
6LA9
 
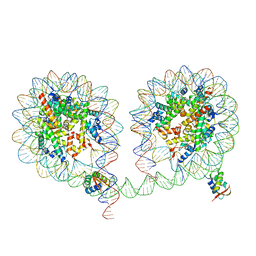 | | 349 bp di-nucleosome harboring cohesive DNA termini assembled with linker histone H1.0 (high cryoprotectant) | | Descriptor: | CALCIUM ION, DNA (349-MER), Histone H1.0, ... | | Authors: | Adhireksan, Z, Sharma, D, Lee, P.L, Davey, C.A. | | Deposit date: | 2019-11-12 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Near-atomic resolution structures of interdigitated nucleosome fibres.
Nat Commun, 11, 2020
|
|
6L9Z
 
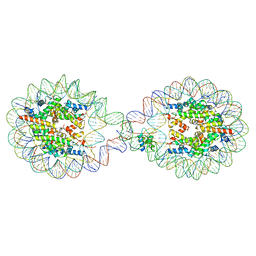 | | 338 bp di-nucleosome assembled with linker histone H1.X | | Descriptor: | CALCIUM ION, CHLORIDE ION, DNA (338-MER), ... | | Authors: | Adhireksan, Z, Sharma, D, Lee, P.L, Davey, C.A. | | Deposit date: | 2019-11-11 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Engineering nucleosomes for generating diverse chromatin assemblies.
Nucleic Acids Res., 49, 2021
|
|
6LAB
 
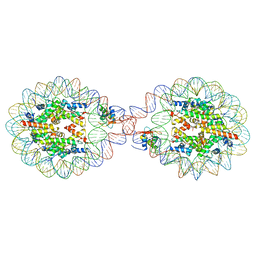 | | 169 bp nucleosome, harboring cohesive DNA termini, assembled with linker histone H1.0 | | Descriptor: | CALCIUM ION, CHLORIDE ION, DNA (169-MER), ... | | Authors: | Adhireksan, Z, Sharma, D, Bao, Q, Lee, P.L, Padavattan, S, Davey, C.A. | | Deposit date: | 2019-11-12 | | Release date: | 2021-02-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Engineering nucleosomes for generating diverse chromatin assemblies.
Nucleic Acids Res., 49, 2021
|
|
6LA2
 
 | | 343 bp di-nucleosome harboring cohesive DNA termini assembled with linker histone H1.0 | | Descriptor: | DNA (343-MER), Histone H1.0, Histone H2A type 1-B/E, ... | | Authors: | Adhireksan, Z, Sharma, D, Lee, P.L, Davey, C.A. | | Deposit date: | 2019-11-11 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.89 Å) | | Cite: | Engineering nucleosomes for generating diverse chromatin assemblies.
Nucleic Acids Res., 49, 2021
|
|
6LER
 
 | | 169 bp nucleosome harboring non-identical cohesive DNA termini. | | Descriptor: | CALCIUM ION, DNA (169-MER), Histone H2A type 1-B/E, ... | | Authors: | Sharma, D, Adhireksan, Z, Lee, P.L, Davey, C.A. | | Deposit date: | 2019-11-26 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Engineering nucleosomes for generating diverse chromatin assemblies.
Nucleic Acids Res., 49, 2021
|
|
1QIQ
 
 | | ISOPENICILLIN N SYNTHASE FROM ASPERGILLUS NIDULANS (ACmC Fe COMPLEX) | | Descriptor: | FE (III) ION, ISOPENICILLIN N SYNTHASE, N-[N-[2-AMINO-6-OXO-HEXANOIC ACID-6-YL]CYSTEINYL]-S-METHYLCYSTEINE, ... | | Authors: | Rutledge, P.J, Clifton, I.J, Burzlaff, N.I, Roach, P.L, Adlington, R.M, Baldwin, J.E. | | Deposit date: | 1999-06-15 | | Release date: | 2000-06-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Reaction Cycle of Isopenicillin N Synthase Observed by X-Ray Diffraction.
Nature, 401, 1999
|
|
1QJF
 
 | | ISOPENICILLIN N SYNTHASE FROM ASPERGILLUS NIDULANS (Monocyclic Sulfoxide - Fe COMPLEX) | | Descriptor: | 1-[(1S)-CARBOXY-2-(METHYLSULFINYL)ETHYL]-(3R)-[(5S)-5-AMINO-5-CARBOXYPENTANAMIDO]-(4R)-SULFANYLAZETIDIN-2-ONE, FE (II) ION, ISOPENICILLIN N SYNTHASE, ... | | Authors: | Rutledge, P.J, Clifton, I.J, Burzlaff, N.I, Roach, P.L, Adlington, R.M, Baldwin, J.E. | | Deposit date: | 1999-06-23 | | Release date: | 2000-06-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Reaction Cycle of Isopenicillin N Synthase Observed by X-Ray Diffraction
Nature, 401, 1999
|
|
1QJE
 
 | | Isopenicillin N synthase from Aspergillus nidulans (IP1 - Fe complex) | | Descriptor: | FE (II) ION, ISOPENICILLIN N, ISOPENICILLIN N SYNTHASE, ... | | Authors: | Burzlaff, N.I, Clifton, I.J, Rutledge, P.J, Roach, P.L, Adlington, R.M, Baldwin, J.E. | | Deposit date: | 1999-06-23 | | Release date: | 2000-06-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Reaction Cycle of Isopenicillin N Synthase Observed by X-Ray Diffraction
Nature, 401, 1999
|
|
1R5S
 
 | | Connexin 43 Carboxyl Terminal Domain | | Descriptor: | Gap junction alpha-1 protein | | Authors: | Sorgen, P.L, Duffy, H.S, Mario, D, Sahoo, P, Coombs, W, Delmar, M, Spray, D.C. | | Deposit date: | 2003-10-13 | | Release date: | 2004-10-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural changes in the carboxyl terminus of the gap junction protein connexin43 indicates signaling between binding domains for c-Src and zonula occludens-1
J.Biol.Chem., 279, 2004
|
|
