3E4B
 
 | | Crystal structure of AlgK from Pseudomonas fluorescens WCS374r | | 分子名称: | AlgK, CHLORIDE ION, GLYCEROL | | 著者 | Keiski, C.-L, Harwich, M, Jain, S, Neculai, A.M, Yip, P, Robinson, H, Whitney, J.C, Burrows, L.L, Ohman, D.E, Howell, P.L. | | 登録日 | 2008-08-11 | | 公開日 | 2009-08-25 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | AlgK is a TPR-containing protein and the periplasmic component of a novel exopolysaccharide secretin.
Structure, 18, 2010
|
|
3FRL
 
 | | The 2.25 A crystal structure of LipL32, the major surface antigen of Leptospira interrogans serovar Copenhageni | | 分子名称: | 2,2',2''-NITRILOTRIETHANOL, CHLORIDE ION, LipL32, ... | | 著者 | Farah, C.S, Guzzo, C.R, Hauk, P, Ho, P.L. | | 登録日 | 2009-01-08 | | 公開日 | 2009-06-16 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structure and calcium-binding activity of LipL32, the major surface antigen of pathogenic Leptospira sp.
J.Mol.Biol., 390, 2009
|
|
3FZ2
 
 | | Crystal structure of the tail terminator protein from phage lambda (gpU-D74A) | | 分子名称: | Minor tail protein U, SULFATE ION | | 著者 | Pell, L.G, Liu, A, Edmonds, E, Donaldson, L.W, Howell, P.L, Davidson, A.R. | | 登録日 | 2009-01-23 | | 公開日 | 2009-05-26 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | The X-ray crystal structure of the phage lambda tail terminator protein reveals the biologically relevant hexameric ring structure and demonstrates a conserved mechanism of tail termination among diverse long-tailed phages.
J.Mol.Biol., 389, 2009
|
|
2AEO
 
 | | Crystal structure of cisplatinated bovine Cu,Zn superoxide dismutase | | 分子名称: | COPPER (II) ION, Cisplatin, Superoxide dismutase [Cu-Zn], ... | | 著者 | Calderone, V, Casini, A, Mangani, S, Messori, L, Orioli, P.L. | | 登録日 | 2005-07-23 | | 公開日 | 2006-05-02 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural investigation of cisplatin-protein interactions: selective platination of His19 in a cuprozinc superoxide dismutase.
Angew. Chem. Int. Ed. Engl., 45, 2006
|
|
5BX9
 
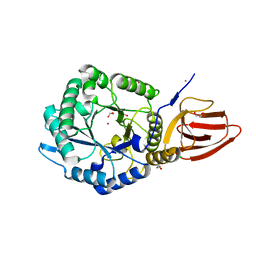 | | Structure of PslG from Pseudomonas aeruginosa | | 分子名称: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | 著者 | Baker, P, Little, D.J, Howell, P.L. | | 登録日 | 2015-06-08 | | 公開日 | 2015-10-07 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.001 Å) | | 主引用文献 | Characterization of the Pseudomonas aeruginosa Glycoside Hydrolase PslG Reveals That Its Levels Are Critical for Psl Polysaccharide Biosynthesis and Biofilm Formation.
J.Biol.Chem., 290, 2015
|
|
6AU1
 
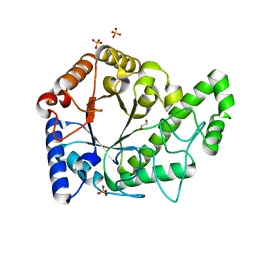 | | Structure of the PgaB (BpsB) glycoside hydrolase domain from Bordetella bronchiseptica | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Putative hemin storage protein, ... | | 著者 | Little, D.J, Bamford, N.C, Howell, P.L. | | 登録日 | 2017-08-30 | | 公開日 | 2018-04-18 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | PgaB orthologues contain a glycoside hydrolase domain that cleaves deacetylated poly-beta (1,6)-N-acetylglucosamine and can disrupt bacterial biofilms.
PLoS Pathog., 14, 2018
|
|
6M8M
 
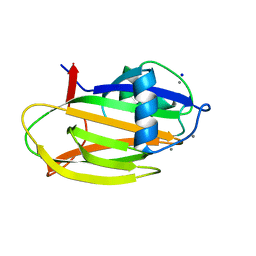 | | PA14 sugar-binding domain from RTX adhesin | | 分子名称: | CALCIUM ION, Putative large adhesion protein (Lap) involved in biofilm formation, SODIUM ION, ... | | 著者 | Vance, T.D.R, Conroy, B, Davies, P.L. | | 登録日 | 2018-08-22 | | 公開日 | 2019-09-04 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Structure and functional analysis of a bacterial adhesin sugar-binding domain.
Plos One, 14, 2019
|
|
6BDT
 
 | |
5C7R
 
 | | Revealing surface waters on an antifreeze protein by fusion protein crystallography | | 分子名称: | Fusion protein of Maltose-binding periplasmic protein and Type-3 ice-structuring protein HPLC 12, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | 著者 | Sun, T, Gauthier, S, Campbell, R.L, Davies, P.L. | | 登録日 | 2015-06-24 | | 公開日 | 2015-09-30 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Revealing Surface Waters on an Antifreeze Protein by Fusion Protein Crystallography Combined with Molecular Dynamic Simulations.
J.Phys.Chem.B, 119, 2015
|
|
7XX6
 
 | | Crystal Structure of Nucleosome-H1.0 Linker Histone Assembly (sticky-169a DNA fragment) | | 分子名称: | CALCIUM ION, DNA (169-MER), Histone H1.0, ... | | 著者 | Adhireksan, Z, Qiuye, B, Lee, P.L, Sharma, D, Padavattan, S, Davey, C.A. | | 登録日 | 2022-05-28 | | 公開日 | 2023-05-31 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.39 Å) | | 主引用文献 | Crystal Structure of Nucleosome-H1.0 Linker Histone Assembly (sticky-169a DNA fragment)
To Be Published
|
|
7XVM
 
 | | Crystal Structure of Nucleosome-H5 Linker Histone Assembly (sticky-169a DNA fragment) | | 分子名称: | CALCIUM ION, CHLORIDE ION, DNA (169-MER), ... | | 著者 | Adhireksan, Z, Qiuye, B, Lee, P.L, Sharma, D, Padavattan, S, Davey, C.A. | | 登録日 | 2022-05-24 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.84 Å) | | 主引用文献 | Crystal Structure of Nucleosome-H1.0 Linker Histone Assembly (sticky-169a DNA fragment)
To Be Published
|
|
7XVL
 
 | | Crystal Structure of Nucleosome-H1.0 Linker Histone Assembly (sticky-169an DNA fragment) | | 分子名称: | DNA (169-MER), Histone H1.0, Histone H2A type 1-B/E, ... | | 著者 | Adhireksan, Z, Qiuye, B, Lee, P.L, Sharma, D, Padavattan, S, Davey, C.A. | | 登録日 | 2022-05-24 | | 公開日 | 2023-05-24 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.506 Å) | | 主引用文献 | Crystal Structure of Nucleosome-H1.0 Linker Histone Assembly (sticky-169an DNA fragment)
To Be Published
|
|
5C5G
 
 | |
7XX5
 
 | | Crystal Structure of Nucleosome-H1.3 Linker Histone Assembly (sticky-169a DNA fragment) | | 分子名称: | CALCIUM ION, DNA (169-MER), Histone H1.3, ... | | 著者 | Adhireksan, Z, Qiuye, B, Lee, P.L, Sharma, D, Padavattan, S, Davey, C.A. | | 登録日 | 2022-05-28 | | 公開日 | 2023-05-31 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.19 Å) | | 主引用文献 | Crystal Structure of Nucleosome-H1.0 Linker Histone Assembly (sticky-169a DNA fragment)
To Be Published
|
|
6BG8
 
 | |
6BGP
 
 | |
6BJD
 
 | | Crystal Structure of Human Calpain-3 Protease Core in Complex with E-64 | | 分子名称: | CALCIUM ION, CHLORIDE ION, Calpain-3, ... | | 著者 | Ye, Q, Campbell, R.L, Davies, P.L. | | 登録日 | 2017-11-06 | | 公開日 | 2018-02-07 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structures of human calpain-3 protease core with and without bound inhibitor reveal mechanisms of calpain activation.
J. Biol. Chem., 293, 2018
|
|
6BKJ
 
 | |
6BT4
 
 | | Crystal structure of the SLH domain of Sap from Bacillus anthracis in complex with a pyruvylated SCWP unit | | 分子名称: | 2-(acetylamino)-4-O-{2-(acetylamino)-4,6-O-[(1S)-1-carboxyethylidene]-2-deoxy-beta-D-mannopyranosyl}-2-deoxy-beta-D-glucopyranose, S-layer protein sap, SULFATE ION | | 著者 | Sychantha, D, Chapman, R.N, Bamford, N.C, Boons, G.J, Howell, P.L, Clarke, A.J. | | 登録日 | 2017-12-05 | | 公開日 | 2018-03-21 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.306 Å) | | 主引用文献 | Molecular Basis for the Attachment of S-Layer Proteins to the Cell Wall of Bacillus anthracis.
Biochemistry, 57, 2018
|
|
5CFB
 
 | | Crystal Structure of Human Glycine Receptor alpha-3 Bound to Strychnine | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycine receptor subunit alpha-3,Glycine receptor subunit alpha-3, ... | | 著者 | Shaffer, P.L, Huang, X, Chen, H. | | 登録日 | 2015-07-08 | | 公開日 | 2015-09-30 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.04 Å) | | 主引用文献 | Crystal structure of human glycine receptor-alpha 3 bound to antagonist strychnine.
Nature, 526, 2015
|
|
6CZT
 
 | |
5E79
 
 | | Macromolecular diffractive imaging using imperfect crystals | | 分子名称: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | 著者 | Ayyer, K, Yefanov, O, Oberthur, D, Roy-Chowdhury, S, Galli, L, Mariani, V, Basu, S, Coe, J, Conrad, C.E, Fromme, R, Schaffer, A, Dorner, K, James, D, Kupitz, C, Metz, M, Nelson, G, Xavier, P.L, Beyerlein, K.R, Schmidt, M, Sarrou, I, Spence, J.C.H, Weierstall, U, White, T.A, Yang, J.-H, Zhao, Y, Liang, M, Aquila, A, Hunter, M.S, Koglin, J.E, Boutet, S, Fromme, P, Barty, A, Chapman, H.N. | | 登録日 | 2015-10-12 | | 公開日 | 2017-02-08 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Macromolecular diffractive imaging using imperfect crystals.
Nature, 530, 2016
|
|
6NWA
 
 | | The structure of the photosystem I IsiA super-complex | | 分子名称: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | 著者 | Toporik, H, Li, J, Williams, D, Chiu, P.L, Mazor, Y. | | 登録日 | 2019-02-06 | | 公開日 | 2019-05-29 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.48 Å) | | 主引用文献 | The structure of the stress-induced photosystem I-IsiA antenna supercomplex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
8A5T
 
 | |
8AME
 
 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 N14SA16H | | 分子名称: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | 著者 | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | 登録日 | 1999-01-24 | | 公開日 | 1999-04-29 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
