7FVK
 
 | | PanDDA analysis group deposition -- PHIP in complex with Z409964562 | | Descriptor: | 4-(furan-2-carbonyl)-N-(2-methoxy-5-methylphenyl)piperazine-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Tomlinson, C, Bradshaw, W.J, Koekemoer, L, Krojer, T, Fearon, D, Biggin, P.C, von Delft, F. | | Deposit date: | 2023-03-09 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7DKG
 
 | | Influenza H5N1 nucleoprotein (truncated) in complex with nucleotides | | Descriptor: | Nucleoprotein, RNA (5'-R(P*(OMU)P*(OMU)P*(OMU))-3') | | Authors: | Tang, Y.S, Xu, S, Chen, Y.W, Wang, J.H, Shaw, P.C. | | Deposit date: | 2020-11-24 | | Release date: | 2021-04-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of influenza nucleoprotein complexed with nucleic acid provide insights into the mechanism of RNA interaction.
Nucleic Acids Res., 49, 2021
|
|
3TJ0
 
 | | Crystal Structure of Influenza B Virus Nucleoprotein | | Descriptor: | Nucleoprotein | | Authors: | Ng, A.K.L, Zhang, H, Liu, J, Au, S.W.N, Wang, J, Shaw, P.C. | | Deposit date: | 2011-08-23 | | Release date: | 2012-06-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.233 Å) | | Cite: | Structural basis for RNA binding and homo-oligomer formation by influenza B virus nucleoprotein
J.Virol., 86, 2012
|
|
3LKR
 
 | | Crystal Structure of HLA B*3501 in complex with influenza NP418 epitope from 2009 H1N1 swine origin strain | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-35 alpha chain, ... | | Authors: | Gras, S, Kedzierski, L, Valkenburg, S.A, Liu, Y.C, Denholm, J, Richards, M, Rimmelzwaan, G.F, Doherty, P.C, Turner, S.J, Rossjohn, J, Kedzierska, K. | | Deposit date: | 2010-01-27 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cross-reactive CD8+ T-cell immunity between the pandemic H1N1-2009 and H1N1-1918 influenza A viruses.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3TTT
 
 | | Structure of F413Y variant of E. coli KatE | | Descriptor: | Catalase HPII, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Loewen, P.C, Jha, V. | | Deposit date: | 2011-09-15 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Mutation of Phe413 to Tyr in catalase KatE from Escherichia coli leads to side chain damage and main chain cleavage.
Arch.Biochem.Biophys., 525, 2012
|
|
3LKQ
 
 | | Crystal Structure of HLA B*3501 in complex with influenza NP418 epitope from 1977 strain | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-35 alpha chain, ... | | Authors: | Gras, S, Kedzierski, L, Valkenburg, S.A, Liu, Y.C, Denholm, J, Richards, M, Rimmelzwaan, G.F, Doherty, P.C, Turner, S.J, Rossjohn, J, Kedzierska, K. | | Deposit date: | 2010-01-27 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cross-reactive CD8+ T-cell immunity between the pandemic H1N1-2009 and H1N1-1918 influenza A viruses.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3LKN
 
 | | Crystal Structure of HLA B*3501 in complex with influenza NP418 epitope from 1918 strain | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-35 alpha chain, ... | | Authors: | Gras, S, Kedzierski, L, Valkenburg, S.A, Liu, Y.C, Denholm, J, Richards, M, Rimmelzwaan, G.F, Doherty, P.C, Turner, S.J, Rossjohn, J, Kedzierska, K. | | Deposit date: | 2010-01-27 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cross-reactive CD8+ T-cell immunity between the pandemic H1N1-2009 and H1N1-1918 influenza A viruses.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3U23
 
 | | Atomic resolution crystal structure of the 2nd SH3 domain from human CD2AP (CMS) in complex with a proline-rich peptide from human RIN3 | | Descriptor: | 1,2-ETHANEDIOL, CD2-associated protein, Ras and Rab interactor 3 | | Authors: | Simister, P.C, Rouka, E, Janning, M, Muniz, J.R.C, Kirsch, K.H, Knapp, S, von Delft, F, Filippakopoulos, P, Arrowsmith, C.H, Krojer, T, Edwards, A.M, Weigelt, J, Bountra, C, Feller, S.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-09-30 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Differential Recognition Preferences of the Three Src Homology 3 (SH3) Domains from the Adaptor CD2-associated Protein (CD2AP) and Direct Association with Ras and Rab Interactor 3 (RIN3).
J.Biol.Chem., 290, 2015
|
|
3LTL
 
 | |
3LKO
 
 | | Crystal Structure of HLA B*3501 in complex with influenza NP418 epitope from 1934 strain | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-35 alpha chain, ... | | Authors: | Gras, S, Kedzierski, L, Valkenburg, S.A, Liu, Y.C, Denholm, J, Richards, M, Rimmelzwaan, G.F, Doherty, P.C, Turner, S.J, Rossjohn, J, Kedzierska, K. | | Deposit date: | 2010-01-27 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cross-reactive CD8+ T-cell immunity between the pandemic H1N1-2009 and H1N1-1918 influenza A viruses.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3TTV
 
 | | Structure of the F413E variant of E. coli KatE | | Descriptor: | Catalase HPII, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Loewen, P.C, Jha, V. | | Deposit date: | 2011-09-15 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Mutation of Phe413 to Tyr in catalase KatE from Escherichia coli leads to side chain damage and main chain cleavage.
Arch.Biochem.Biophys., 525, 2012
|
|
3P9Q
 
 | | Structure of I274C variant of E. coli KatE | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE 17R, 18S, ... | | Authors: | Loewen, P.C, Jha, V, Louis, S, Chelikani, P, Carpena, X, Fita, I. | | Deposit date: | 2010-10-18 | | Release date: | 2010-12-22 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Modulation of heme orientation and binding by a single residue in catalase HPII of Escherichia coli.
Biochemistry, 50, 2011
|
|
3TTU
 
 | | Structure of F413Y/H128N double variant of E. coli KatE | | Descriptor: | Catalase HPII, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Loewen, P.C, Jha, V. | | Deposit date: | 2011-09-15 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Mutation of Phe413 to Tyr in catalase KatE from Escherichia coli leads to side chain damage and main chain cleavage.
Arch.Biochem.Biophys., 525, 2012
|
|
3P9S
 
 | | Structure of I274A variant of E. coli KatE | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE 17R, 18S, ... | | Authors: | Loewen, P.C, Jha, V, Louis, S, Chelikani, P, Carpena, X, Fita, I. | | Deposit date: | 2010-10-18 | | Release date: | 2010-12-22 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Modulation of heme orientation and binding by a single residue in catalase HPII of Escherichia coli.
Biochemistry, 50, 2011
|
|
3LKS
 
 | | Crystal Structure of HLA B*3501 in complex with influenza NP418 epitope from 1980 strain | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-35 alpha chain, ... | | Authors: | Gras, S, Kedzierski, L, Valkenburg, S.A, Liu, Y.C, Denholm, J, Richards, M, Rimmelzwaan, G.F, Doherty, P.C, Turner, S.J, Rossjohn, J, Kedzierska, K. | | Deposit date: | 2010-01-27 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cross-reactive CD8+ T-cell immunity between the pandemic H1N1-2009 and H1N1-1918 influenza A viruses.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3TTX
 
 | | Structure of the F413K variant of E. coli KatE | | Descriptor: | Catalase HPII, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Loewen, P.C, Jha, V. | | Deposit date: | 2011-09-15 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Mutation of Phe413 to Tyr in catalase KatE from Escherichia coli leads to side chain damage and main chain cleavage.
Arch.Biochem.Biophys., 525, 2012
|
|
3LKP
 
 | | Crystal Structure of HLA B*3501 in complex with influenza NP418 epitope from 1972 strain | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-35 alpha chain, ... | | Authors: | Gras, S, Kedzierski, L, Valkenburg, S.A, Liu, Y.C, Denholm, J, Richards, M, Rimmelzwaan, G.F, Doherty, P.C, Turner, S.J, Rossjohn, J, Kedzierska, K. | | Deposit date: | 2010-01-27 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cross-reactive CD8+ T-cell immunity between the pandemic H1N1-2009 and H1N1-1918 influenza A viruses.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3P6A
 
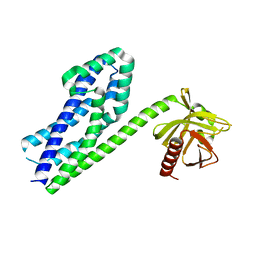 | |
3PQ8
 
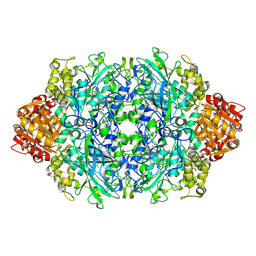 | | Structure of I274C variant of E. coli KatE[] - Images 37-42 | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE 17R, 18S, ... | | Authors: | Loewen, P.C, Jha, V, Louis, S, Chelikani, P, Carpena, X, Fita, I. | | Deposit date: | 2010-11-25 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modulation of heme orientation and binding by a single residue in catalase HPII of Escherichia coli.
Biochemistry, 50, 2011
|
|
3PQ4
 
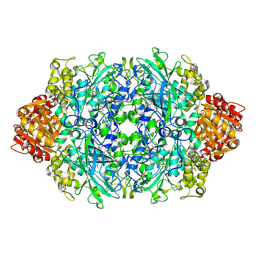 | | Structure of I274C variant of E. coli KatE[] - Images 13-18 | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE 17R, 18S, ... | | Authors: | Loewen, P.C, Jha, V, Louis, S, Chelikani, P, Carpena, X, Fita, I. | | Deposit date: | 2010-11-25 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Modulation of heme orientation and binding by a single residue in catalase HPII of Escherichia coli.
Biochemistry, 50, 2011
|
|
3PQ2
 
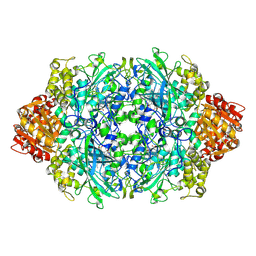 | | Structure of I274C variant of E. coli KatE[] - Images 1-6 | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE 17R, 18S, ... | | Authors: | Loewen, P.C, Jha, V, Louis, S, Chelikani, P, Carpena, X, Fita, I. | | Deposit date: | 2010-11-25 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Modulation of heme orientation and binding by a single residue in catalase HPII of Escherichia coli.
Biochemistry, 50, 2011
|
|
3PIN
 
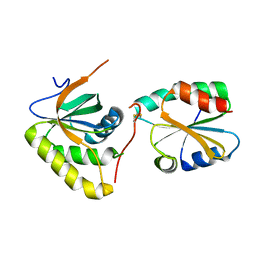 | | Crystal structure of Mxr1 from Saccharomyces cerevisiae in complex with Trx2 | | Descriptor: | Peptide methionine sulfoxide reductase, Thioredoxin-2 | | Authors: | Ma, X.X, Guo, P.C, Shi, W.W, Luo, M, Tan, X.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2010-11-07 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural plasticity of the thioredoxin recognition site of yeast methionine S-sulfoxide reductase Mxr1
J.Biol.Chem., 286, 2011
|
|
3PQ7
 
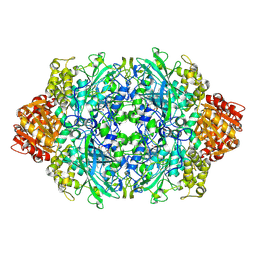 | | Structure of I274C variant of E. coli KatE[] - Images 31-36 | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE 17R, 18S, ... | | Authors: | Loewen, P.C, Jha, V, Louis, S, Chelikani, P, Carpena, X, Fita, I. | | Deposit date: | 2010-11-25 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modulation of heme orientation and binding by a single residue in catalase HPII of Escherichia coli.
Biochemistry, 50, 2011
|
|
3PQ6
 
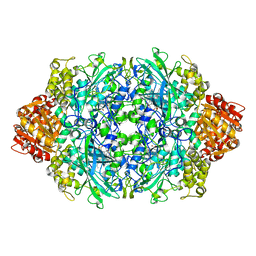 | | Structure of I274C variant of E. coli KatE[] - Images 25-30 | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE 17R, 18S, ... | | Authors: | Loewen, P.C, Jha, V, Louis, S, Chelikani, P, Carpena, X, Fita, I. | | Deposit date: | 2010-11-25 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modulation of heme orientation and binding by a single residue in catalase HPII of Escherichia coli.
Biochemistry, 50, 2011
|
|
3PIM
 
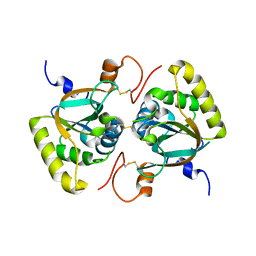 | | Crystal structure of Mxr1 from Saccharomyces cerevisiae in unusual oxidized form | | Descriptor: | Peptide methionine sulfoxide reductase | | Authors: | Ma, X.X, Guo, P.C, Shi, W.W, Luo, M, Tan, X.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2010-11-07 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural plasticity of the thioredoxin recognition site of yeast methionine S-sulfoxide reductase Mxr1
J.Biol.Chem., 286, 2011
|
|
