2VEX
 
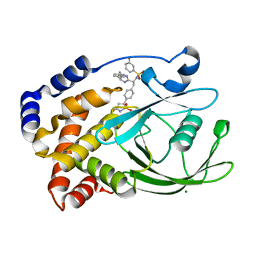 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B IN COMPLEX WITH AN ISOTHIAZOLIDINONE-CONTAINING INHIBITOR | | Descriptor: | MAGNESIUM ION, N-{(1S)-2-{4-[(5S)-1,1-dioxido-3-oxoisothiazolidin-5-yl]phenyl}-1-[(4R)-4-(2-phenylethyl)-4,5-dihydro-1H-imidazol-2-yl]ethyl}-3-fluorobenzenesulfonamide, TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 1 | | Authors: | Douty, B, Wayland, B, Ala, P.J, Bower, M.J, Pruitt, J, Bostrom, L, Wei, M, Klabe, R, Gonneville, L, Wynn, R, Burn, T.C, Liu, P.C.C, Combs, A.P, Yue, E.W. | | Deposit date: | 2007-10-27 | | Release date: | 2007-11-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Isothiazolidinone Inhibitors of Ptp1B Containing Imidazoles and Imidazolines
Bioorg.Med.Chem.Lett., 18, 2008
|
|
1KN3
 
 | |
1ADU
 
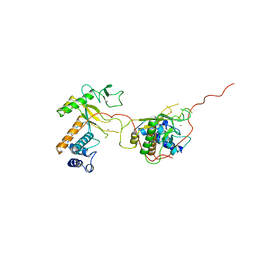 | | EARLY E2A DNA-BINDING PROTEIN | | Descriptor: | ADENOVIRUS SINGLE-STRANDED DNA-BINDING PROTEIN, ZINC ION | | Authors: | Tucker, P.A, Kanellopoulos, P.N, Tsernoglou, D, Van Der Vliet, P.C. | | Deposit date: | 1995-05-11 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Alternative arrangements of the protein chain are possible for the adenovirus single-stranded DNA binding protein.
J.Mol.Biol., 257, 1996
|
|
1A3T
 
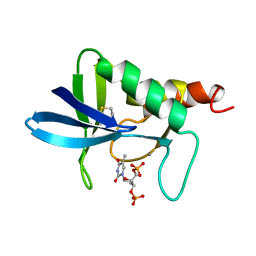 | | STAPHYLOCOCCAL NUCLEASE, V23C VARIANT, COMPLEX WITH 2-FLUOROETHANE THIOL AND 3',5'-THYMIDINE DIPHOSPHATE | | Descriptor: | CALCIUM ION, STAPHYLOCOCCAL NUCLEASE, THYMIDINE-3',5'-DIPHOSPHATE | | Authors: | Wynn, R, Harkins, P.C, Richards, F.M, Fox, R.O. | | Deposit date: | 1998-01-24 | | Release date: | 1998-04-29 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Comparison of straight chain and cyclic unnatural amino acids embedded in the core of staphylococcal nuclease.
Protein Sci., 6, 1997
|
|
2VCN
 
 | | Structure of isoniazid (INH) bound to cytosolic soybean ascorbate peroxidase mutant W41A | | Descriptor: | 4-(DIAZENYLCARBONYL)PYRIDINE, ASCORBATE PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Metcalfe, C.L, Macdonald, I.K, Brown, K.A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2007-09-25 | | Release date: | 2007-12-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The Tuberculosis Prodrug Isoniazid Bound to Activating Peroxidases.
J.Biol.Chem., 283, 2008
|
|
2V2E
 
 | | Structure of isoniazid (INH) bound to cytochrome c peroxidase mutant N184R Y36A | | Descriptor: | 4-(DIAZENYLCARBONYL)PYRIDINE, CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Metcalfe, C.L, Macdonald, I.K, Brown, K.A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2007-06-05 | | Release date: | 2007-12-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The Tuberculosis Prodrug Isoniazid Bound to Activating Peroxidases.
J.Biol.Chem., 283, 2008
|
|
2X08
 
 | | cytochrome c peroxidase: ascorbate bound to the engineered ascorbate binding site | | Descriptor: | ASCORBIC ACID, CYTOCHROME C PEROXIDASE, MITOCHONDRIAL, ... | | Authors: | Murphy, E.J, Metcalfe, C.L, Gumiero, A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2009-12-07 | | Release date: | 2010-11-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Engineering the substrate specificity and reactivity of a heme protein: creation of an ascorbate binding site in cytochrome c peroxidase.
Biochemistry, 47, 2008
|
|
1KV8
 
 | | Crystal Structure of 3-Keto-L-Gulonate 6-Phosphate Decarboxylase | | Descriptor: | 3-Keto-L-Gulonate 6-Phosphate Decarboxylase, MAGNESIUM ION, PHOSPHATE ION | | Authors: | Wise, E, Yew, W.S, Babbitt, P.C, Gerlt, J.A, Rayment, I. | | Deposit date: | 2002-01-25 | | Release date: | 2002-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Homologous (beta/alpha)8-barrel enzymes that catalyze unrelated reactions: orotidine 5'-monophosphate decarboxylase and 3-keto-L-gulonate 6-phosphate decarboxylase.
Biochemistry, 41, 2002
|
|
1BZU
 
 | |
1BJ8
 
 | | THIRD N-TERMINAL DOMAIN OF GP130, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | GP130 | | Authors: | Kernebeck, T, Pflanz, S, Muller-Newen, G, Kurapkat, G, Scheek, R.M, Dijkstra, K, Heinrich, P.C, Wollmer, A, Grzesiek, S, Grotzinger, J. | | Deposit date: | 1998-07-02 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The signal transducer gp130: solution structure of the carboxy-terminal domain of the cytokine receptor homology region.
Protein Sci., 8, 1999
|
|
1BZ3
 
 | |
1BKH
 
 | | MUCONATE LACTONIZING ENZYME FROM PSEUDOMONAS PUTIDA | | Descriptor: | MUCONATE LACTONIZING ENZYME | | Authors: | Hasson, M.S, Schlichting, I, Moulai, J, Taylor, K, Barrett, W, Kenyon, G.L, Babbitt, P.C, Gerlt, J.A, Petsko, G.A, Ringe, D. | | Deposit date: | 1998-07-07 | | Release date: | 1998-10-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Evolution of an enzyme active site: the structure of a new crystal form of muconate lactonizing enzyme compared with mandelate racemase and enolase.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1BZY
 
 | | HUMAN HGPRTASE WITH TRANSITION STATE INHIBITOR | | Descriptor: | HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE, MAGNESIUM ION, PHOSPHORIC ACID MONO-[5-(2-AMINO-4-OXO-4,5-DIHYDRO-3H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)-3,4-DIHYDROXY-PYRROLIDIN-2-YLMETHYL] ESTER, ... | | Authors: | Shi, W, Li, C, Tyler, P.C, Furneaux, R.H, Grubmeyer, C, Schramm, V.L, Almo, S.C. | | Deposit date: | 1998-11-05 | | Release date: | 1999-06-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0 A structure of human hypoxanthine-guanine phosphoribosyltransferase in complex with a transition-state analog inhibitor.
Nat.Struct.Biol., 6, 1999
|
|
3POB
 
 | |
3POG
 
 | |
3PON
 
 | |
3PQ7
 
 | | Structure of I274C variant of E. coli KatE[] - Images 31-36 | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE 17R, 18S, ... | | Authors: | Loewen, P.C, Jha, V, Louis, S, Chelikani, P, Carpena, X, Fita, I. | | Deposit date: | 2010-11-25 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modulation of heme orientation and binding by a single residue in catalase HPII of Escherichia coli.
Biochemistry, 50, 2011
|
|
2VEU
 
 | | Crystal structure of protein tyrosine phosphatase 1B in complex with an isothiazolidinone-containing inhibitor | | Descriptor: | N-[(1S)-2-{4-[(5S)-1,1-dioxido-3-oxoisothiazolidin-5-yl]phenyl}-1-(4-phenyl-1H-imidazol-2-yl)ethyl]-3-(trifluoromethyl)benzenesulfonamide, TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 1 | | Authors: | Douty, B, Wayland, B, Ala, P.J, Bower, M.J, Pruitt, J, Bostrom, L, Wei, M, Klabe, R, Gonneville, L, Wynn, R, Burn, T.C, Liu, P.C.C, Combs, A.P, Yue, E.W. | | Deposit date: | 2007-10-27 | | Release date: | 2007-11-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Isothiazolidinone Inhibitors of Ptp1B Containing Imidazoles and Imidazolines
Bioorg.Med.Chem.Lett., 18, 2008
|
|
1LR1
 
 | | Solution Structure of the Oligomerization Domain of the Bacterial Chromatin-Structuring Protein H-NS | | Descriptor: | dna-binding protein h-ns | | Authors: | Esposito, D, Petrovic, A, Harris, R, Ono, S, Eccleston, J, Mbabaali, A, Haq, I, Higgins, C.F, Hinton, J.C.D, Driscoll, P.C, Ladbury, J.E. | | Deposit date: | 2002-05-14 | | Release date: | 2003-01-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | H-NS Oligomerization Domain Structure Reveals the Mechanism for High Order
Self-association of the Intact Protein
J.Mol.Biol., 324, 2002
|
|
3PGQ
 
 | | Crystal Structure of the Carboxyltransferase Domain of S. cerevisiae Acetyl CoA Carboxylase in Complex with Pinoxaden | | Descriptor: | 8-(2-ethenyl-6-ethyl-4-methylphenyl)tetrahydro-7H-pyrazolo[1,2-d][1,4,5]oxadiazepine-7,9(8H)-dione, Acetyl-CoA carboxylase | | Authors: | Tong, L, Yu, L.P.C, Kim, Y.S. | | Deposit date: | 2010-11-02 | | Release date: | 2010-12-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanism for the inhibition of the carboxyltransferase domain of acetyl-coenzyme A carboxylase by pinoxaden.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3PIM
 
 | | Crystal structure of Mxr1 from Saccharomyces cerevisiae in unusual oxidized form | | Descriptor: | Peptide methionine sulfoxide reductase | | Authors: | Ma, X.X, Guo, P.C, Shi, W.W, Luo, M, Tan, X.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2010-11-07 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural plasticity of the thioredoxin recognition site of yeast methionine S-sulfoxide reductase Mxr1
J.Biol.Chem., 286, 2011
|
|
3POE
 
 | |
3PQ6
 
 | | Structure of I274C variant of E. coli KatE[] - Images 25-30 | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE 17R, 18S, ... | | Authors: | Loewen, P.C, Jha, V, Louis, S, Chelikani, P, Carpena, X, Fita, I. | | Deposit date: | 2010-11-25 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modulation of heme orientation and binding by a single residue in catalase HPII of Escherichia coli.
Biochemistry, 50, 2011
|
|
1CZ1
 
 | | EXO-B-(1,3)-GLUCANASE FROM CANDIDA ALBICANS AT 1.85 A RESOLUTION | | Descriptor: | PROTEIN (EXO-B-(1,3)-GLUCANASE) | | Authors: | Cutfield, S.M, Davies, G.J, Murshudov, G, Anderson, B.F, Moody, P.C.E, Sullivan, P.A, Cutfield, J.F. | | Deposit date: | 1999-09-01 | | Release date: | 2000-01-03 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structure of the exo-beta-(1,3)-glucanase from Candida albicans in native and bound forms: relationship between a pocket and groove in family 5 glycosyl hydrolases.
J.Mol.Biol., 294, 1999
|
|
1K89
 
 | | K89L MUTANT OF GLUTAMATE DEHYDROGENASE | | Descriptor: | GLUTAMATE DEHYDROGENASE | | Authors: | Stillman, T.J, Migueis, A.M.B, Wang, X.G, Baker, P.J, Britton, K.L, Engel, P.C, Rice, D.W. | | Deposit date: | 1998-06-05 | | Release date: | 1999-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Insights into the mechanism of domain closure and substrate specificity of glutamate dehydrogenase from Clostridium symbiosum.
J.Mol.Biol., 285, 1999
|
|
