8DT3
 
 | | Cryo-EM structure of spike binding to Fab of neutralizing antibody (locally refined) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain Fab of SW186, Light chain Fab of SW186, ... | | Authors: | Sun, P.C, Fang, Y, Bai, X.C, Chen, Z.J. | | Deposit date: | 2022-07-25 | | Release date: | 2022-08-03 | | Last modified: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | An antibody that neutralizes SARS-CoV-1 and SARS-CoV-2 by binding to a conserved spike epitope outside the receptor binding motif.
Sci Immunol, 7, 2022
|
|
1M6H
 
 | | Human glutathione-dependent formaldehyde dehydrogenase | | Descriptor: | Glutathione-dependent formaldehyde dehydrogenase, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Sanghani, P.C, Robinson, H, Bosron, W.F, Hurley, T.D. | | Deposit date: | 2002-07-16 | | Release date: | 2002-07-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human glutathione-dependent formaldehyde dehydrogenase. Structures of apo, binary, and inhibitory ternary complexes.
Biochemistry, 41, 2002
|
|
1MA0
 
 | | Ternary complex of Human glutathione-dependent formaldehyde dehydrogenase with NAD+ and dodecanoic acid | | Descriptor: | Glutathione-dependent formaldehyde dehydrogenase, LAURIC ACID, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Sanghani, P.C, Robinson, H, Bosron, W.F, Hurley, T.D. | | Deposit date: | 2002-07-30 | | Release date: | 2002-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Human glutathione-dependent formaldehyde dehydrogenase. Structures of apo, binary, and inhibitory ternary complexes.
Biochemistry, 41, 2002
|
|
1M7S
 
 | | Crystal Structure Analysis of Catalase CatF of Pseudomonas syringae | | Descriptor: | Catalase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Carpena, X, Soriano, M, Klotz, M.G, Duckworth, H.W, Donald, L.J, Melik-Adamyan, W, Fita, I, Loewen, P.C. | | Deposit date: | 2002-07-22 | | Release date: | 2002-08-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Clade 1 catalase, CatF of Pseudomonas syringae, at 1.8 A resolution
Proteins, 50, 2003
|
|
1MP0
 
 | | Binary Complex of Human Glutathione-Dependent Formaldehyde Dehydrogenase with NAD(H) | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Sanghani, P.C, Robinson, H, Hurley, T.D, Bosron, W.F. | | Deposit date: | 2002-09-10 | | Release date: | 2002-09-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-function relationships in human Class III alcohol dehydrogenase
(formaldehyde dehydrogenase)
Chem.Biol.Interact., 143, 2003
|
|
1MC5
 
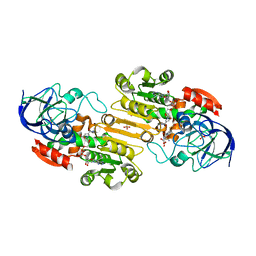 | | Ternary complex of Human glutathione-dependent formaldehyde dehydrogenase with S-(hydroxymethyl)glutathione and NADH | | Descriptor: | 2-AMINO-4-[1-CARBOXYMETHYL-CARBAMOYL)-2-HYDROXYMETHYLSULFANYL-ETHYLCARBAMOYL]-BUTYRIC ACID, Alcohol dehydrogenase class III chi chain, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Sanghani, P.C, Bosron, W.F, Hurley, T.D. | | Deposit date: | 2002-08-05 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Human glutathione-dependent formaldehyde dehydrogenase.
Structural changes associated with Ternary Complex formation
Biochemistry, 41, 2002
|
|
1MDY
 
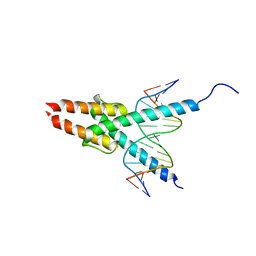 | | CRYSTAL STRUCTURE OF MYOD BHLH DOMAIN BOUND TO DNA: PERSPECTIVES ON DNA RECOGNITION AND IMPLICATIONS FOR TRANSCRIPTIONAL ACTIVATION | | Descriptor: | DNA (5'-D(*TP*CP*AP*AP*CP*AP*GP*CP*TP*GP*TP*TP*GP*A)-3'), PROTEIN (MYOD BHLH DOMAIN) | | Authors: | Ma, P.C.M, Rould, M.A, Weintraub, H, Pabo, C.O. | | Deposit date: | 1994-06-09 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of MyoD bHLH domain-DNA complex: perspectives on DNA recognition and implications for transcriptional activation.
Cell(Cambridge,Mass.), 77, 1994
|
|
1MBL
 
 | |
8FWY
 
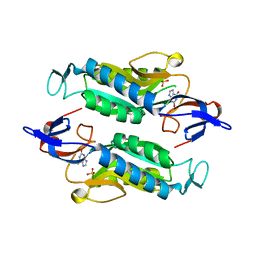 | | Crystal structure of the Trypanosoma cruzi hypoxanthine-guanine-xanthine phosphoribosyltransferase (HGXPRT), isoform D, bound to the dead-end complex xanthine and pyrophosphate | | Descriptor: | Hypoxanthine-guanine phosphoribosyltransferase, PHOSPHATE ION, XANTHINE | | Authors: | Hughes, R, Meneely, K.M, Glockzin, K, Tyler, P.C, Lamb, A.L, Meek, T.D, Katzfuss, A. | | Deposit date: | 2023-01-23 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Kinetic and Structural Characterization of Trypanosoma cruzi Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferases and Repurposing of Transition-State Analogue Inhibitors.
Biochemistry, 62, 2023
|
|
8FX3
 
 | | Crystal structure of the Trypanosoma cruzi hypoxanthine-guanine-xanthine phosphoribosyltransferase (HGXPRT), isoform D, bound to Immucillin-GP, showing the structure of the complete active site in its open conformation | | Descriptor: | Hypoxanthine-guanine phosphoribosyltransferase, PHOSPHORIC ACID MONO-[5-(2-AMINO-4-OXO-4,5-DIHYDRO-3H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)-3,4-DIHYDROXY-PYRROLIDIN-2-YLMETHYL] ESTER | | Authors: | Hughes, R, Meneely, K.M, Glockzin, K, Suthagar, K, Tyler, P.C, Lamb, A.L, Meek, T.D, Katzfuss, A. | | Deposit date: | 2023-01-23 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Kinetic and Structural Characterization of Trypanosoma cruzi Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferases and Repurposing of Transition-State Analogue Inhibitors.
Biochemistry, 62, 2023
|
|
8FX1
 
 | | Crystal structure of the Trypanosoma cruzi hypoxanthine-guanine-xanthine phosphoribosyltransferase (HGXPRT), isoform D, bound to (R)-SerMe-ImmH Phosphonate | | Descriptor: | Hypoxanthine-guanine phosphoribosyltransferase, [(3R)-4-hydroxy-3-{[(4-oxo-4,5-dihydro-3H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]amino}butyl]phosphonic acid | | Authors: | Hughes, R, Meneely, K.M, Glockzin, K, Clinch, K, Tyler, P.C, Lamb, A.L, Meek, T.D, Katzfuss, A. | | Deposit date: | 2023-01-23 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Kinetic and Structural Characterization of Trypanosoma cruzi Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferases and Repurposing of Transition-State Analogue Inhibitors.
Biochemistry, 62, 2023
|
|
8FX0
 
 | | Crystal structure of the Trypanosoma cruzi hypoxanthine-guanine-xanthine phosphoribosyltransferase (HGXPRT), isoform D, bound to (S)-SerMe-ImmH Phosphonate | | Descriptor: | Hypoxanthine-guanine phosphoribosyltransferase, [(3R)-4-hydroxy-3-{[(4-oxo-4,5-dihydro-3H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]amino}butyl]phosphonic acid | | Authors: | Hughes, R, Meneely, K.M, Glockzin, K, Clinch, K, Tyler, P.C, Lamb, A.L, Meek, T.D, Katzfuss, A. | | Deposit date: | 2023-01-23 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Kinetic and Structural Characterization of Trypanosoma cruzi Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferases and Repurposing of Transition-State Analogue Inhibitors.
Biochemistry, 62, 2023
|
|
8FX2
 
 | | Crystal structure of the Trypanosoma cruzi hypoxanthine-guanine-xanthine phosphoribosyltransferase (HGXPRT), isoform D, bound to Immucillin-HP | | Descriptor: | (1S)-1(9-DEAZAHYPOXANTHIN-9YL)1,4-DIDEOXY-1,4-IMINO-D-RIBITOL-5-PHOSPHATE, Hypoxanthine-guanine phosphoribosyltransferase | | Authors: | Hughes, R, Meneely, K.M, Glockzin, K, Suthagar, K, Tyler, P.C, Lamb, A.L, Meek, T.D, Katzfuss, A. | | Deposit date: | 2023-01-23 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Kinetic and Structural Characterization of Trypanosoma cruzi Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferases and Repurposing of Transition-State Analogue Inhibitors.
Biochemistry, 62, 2023
|
|
8FWZ
 
 | | Crystal structure of the Trypanosoma cruzi hypoxanthine-guanine-xanthine phosphoribosyltransferase (HGXPRT), isoform D, bound to Hydroxypropyl-Lin-ImmH Phosphonate | | Descriptor: | Hypoxanthine-guanine phosphoribosyltransferase, PHOSPHATE ION, [(2R)-2-hydroxy-3-{[(4-oxo-4,5-dihydro-3H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]amino}propyl]phosphonic acid | | Authors: | Hughes, R, Meneely, K.M, Glockzin, K, Clinch, K, Tyler, P.C, Lamb, A.L, Meek, T.D, Katzfuss, A. | | Deposit date: | 2023-01-23 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Kinetic and Structural Characterization of Trypanosoma cruzi Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferases and Repurposing of Transition-State Analogue Inhibitors.
Biochemistry, 62, 2023
|
|
6KTH
 
 | | Crystal structure of Juvenile hormone diol kinase JHDK-L2 from silkworm, Bombyx mori | | Descriptor: | CALCIUM ION, GLYCEROL, Juvenile hormone diol kinase | | Authors: | Zhang, Y.S, Xu, H.Y, Wang, Z, Zhang, L, Zhao, P, Guo, P.C. | | Deposit date: | 2019-08-28 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Structural characterization and functional analysis of juvenile hormone diol kinase from the silkworm, Bombyx mori.
Int.J.Biol.Macromol., 167, 2021
|
|
6KSH
 
 | | Crystal structure of pyruvate kinase (PYK) from Plasmodium falciparum in complex with oxalate and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, OXALATE ION, ... | | Authors: | Zhong, W, Cai, Q, Li, K, Lescar, J, Dedon, P.C. | | Deposit date: | 2019-08-23 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Pyruvate kinase from Plasmodium falciparum: Structural and kinetic insights into the allosteric mechanism.
Biochem.Biophys.Res.Commun., 532, 2020
|
|
7CEM
 
 | | Crystal Structure of YbeA CP74 W7F | | Descriptor: | Ribosomal RNA large subunit methyltransferase H,Ribosomal RNA large subunit methyltransferase H | | Authors: | Liu, C.Y, Wu, C.Y, Lai, C.H, Hsu, S.T.D, Lyu, P.C. | | Deposit date: | 2020-06-23 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4251 Å) | | Cite: | Crystal Structure of YbeA CP74 W7F
To Be Published
|
|
6L9K
 
 | | H2-Ld a1a2 complexed with A5 peptide | | Descriptor: | H2-Ld a1a2, SER-PRO-SER-TYR-ALA-TYR-HIS-GLN-PHE | | Authors: | Wei, P.C, Yin, L. | | Deposit date: | 2019-11-10 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures suggest an approach for converting weak self-peptide tumor antigens into superagonists for CD8 T cells in cancer.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6L9O
 
 | | Crystal structure of FABP7 apo | | Descriptor: | Fatty acid-binding protein, brain | | Authors: | Wei, P.C, Yin, L. | | Deposit date: | 2019-11-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal structure of FABP7 apo
To Be Published
|
|
7CF7
 
 | | Crystal Structure of YbeA CP74 W72F | | Descriptor: | Ribosomal RNA large subunit methyltransferase H,Ribosomal RNA large subunit methyltransferase H | | Authors: | Liu, C.Y, Lai, C.H, Hsu, S.T.D, Lyu, P.C. | | Deposit date: | 2020-06-24 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6178 Å) | | Cite: | Crystal Structure of YbeA CP74 W72F
To Be Published
|
|
7CFY
 
 | | Crystal Structure of YbeA CP74 W48F | | Descriptor: | Ribosomal RNA large subunit methyltransferase H,Ribosomal RNA large subunit methyltransferase H | | Authors: | Liu, C.Y, Lai, C.H, Hsu, S.T.D, Lyu, P.C. | | Deposit date: | 2020-06-29 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4051 Å) | | Cite: | Crystal Structure of YbeA CP74 W48F
To Be Published
|
|
7CIX
 
 | |
7CIW
 
 | |
7CIU
 
 | |
6L9N
 
 | | H2-Ld complexed with A5 peptide | | Descriptor: | MHC, SER-PRO-SER-TYR-ALA-TYR-HIS-GLN-PHE, b2m | | Authors: | Wei, P.C, Yin, L. | | Deposit date: | 2019-11-10 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures suggest an approach for converting weak self-peptide tumor antigens into superagonists for CD8 T cells in cancer.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
