1KV8
 
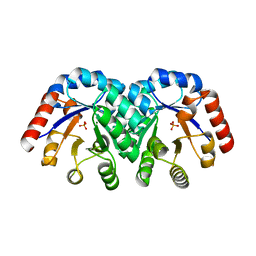 | | Crystal Structure of 3-Keto-L-Gulonate 6-Phosphate Decarboxylase | | Descriptor: | 3-Keto-L-Gulonate 6-Phosphate Decarboxylase, MAGNESIUM ION, PHOSPHATE ION | | Authors: | Wise, E, Yew, W.S, Babbitt, P.C, Gerlt, J.A, Rayment, I. | | Deposit date: | 2002-01-25 | | Release date: | 2002-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Homologous (beta/alpha)8-barrel enzymes that catalyze unrelated reactions: orotidine 5'-monophosphate decarboxylase and 3-keto-L-gulonate 6-phosphate decarboxylase.
Biochemistry, 41, 2002
|
|
1JVK
 
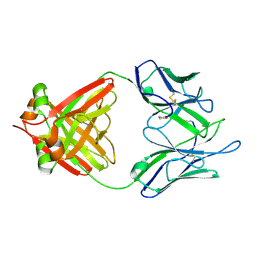 | | THREE-DIMENSIONAL STRUCTURE OF AN IMMUNOGLOBULIN LIGHT CHAIN DIMER ACTING AS A LETHAL AMYLOID PRECURSOR | | Descriptor: | IMMUNOGLOBULIN LAMBDA LIGHT CHAIN | | Authors: | Bourne, P.C, Ramsland, P.A, Shan, L, Fan, Z.-C, DeWitt, C.R, Shultz, B.B, Terzyan, S.S, Edmundson, A.B. | | Deposit date: | 2001-08-30 | | Release date: | 2002-05-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Three-dimensional structure of an immunoglobulin light-chain dimer with amyloidogenic properties.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1QLW
 
 | |
1N3I
 
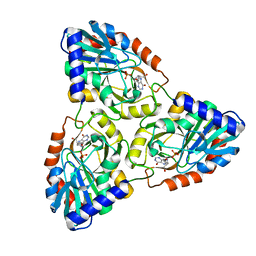 | | Crystal Structure of Mycobacterium tuberculosis PNP with transition state analog DADMe-ImmH | | Descriptor: | 7-[[(3R,4R)-3-(hydroxymethyl)-4-oxidanyl-pyrrolidin-1-ium-1-yl]methyl]-3,5-dihydropyrrolo[3,2-d]pyrimidin-4-one, PHOSPHATE ION, Purine Nucleoside Phosphorylase | | Authors: | Lewandowicz, A, Shi, W, Evans, G.B, Tyler, P.C, Furneaux, R.H, Basso, L.A, Santos, D.S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2002-10-28 | | Release date: | 2003-09-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Over-The-Barrier Transition State Analogues Provide New Chemistries for Inhibitor Design: The Case of Purine Nucleoside Phosphorylase
BIOCHEMISTRY, 42, 2003
|
|
1N4A
 
 | | The Ligand Bound Structure of E.coli BtuF, the Periplasmic Binding Protein for Vitamin B12 | | Descriptor: | CYANOCOBALAMIN, Vitamin B12 transport protein btuF | | Authors: | Karpowich, N.K, Smith, P.C, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-10-30 | | Release date: | 2003-03-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the BtuF periplasmic-binding protein for vitamin B12 suggest a functionally important reduction in protein mobility upon ligand binding.
J.Biol.Chem., 278, 2003
|
|
1OKF
 
 | | NMR structure of an alpha-L-LNA:RNA hybrid | | Descriptor: | 5'-D(*CP*ATLP*GP*AP*ATLP*AP*ATLP*GP*CP)-3', 5'-R(*GP*CP*AP*UP*AP*UP*CP*AP*GP)-3' | | Authors: | Nielsen, J.T, Stein, P.C, Petersen, M. | | Deposit date: | 2003-07-23 | | Release date: | 2003-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of an Alpha-L-Lna:RNA Hybrid: Structural Implications for Rnase H Recognition
Nucleic Acids Res., 31, 2003
|
|
1MBL
 
 | |
1MPJ
 
 | | X-RAY CRYSTALLOGRAPHIC STUDIES ON HEXAMERIC INSULINS IN THE PRESENCE OF HELIX-STABILIZING AGENTS, THIOCYANATE, METHYLPARABEN AND PHENOL | | Descriptor: | CHLORIDE ION, PHENOL, PHENOL INSULIN, ... | | Authors: | Whittingham, J.L, Dodson, E.J, Moody, P.C.E, Dodson, G.G. | | Deposit date: | 1995-09-13 | | Release date: | 1996-01-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray crystallographic studies on hexameric insulins in the presence of helix-stabilizing agents, thiocyanate, methylparaben, and phenol.
Biochemistry, 34, 1995
|
|
1PNK
 
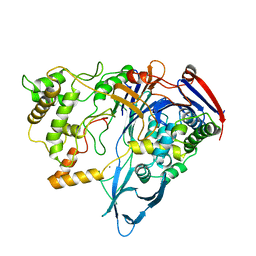 | |
1MZH
 
 | | QR15, an Aldolase | | Descriptor: | Deoxyribose-phosphate aldolase, PHOSPHATE ION | | Authors: | Tan, A.Y, Smith, P.C, Shen, J, Xiao, R, Acton, T, Rost, B, Montelione, G, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-10-07 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Aquifex Aeolicus Aldolase,
Northeast Structural Genomics Consortium Target
QR15
To be Published
|
|
1PNJ
 
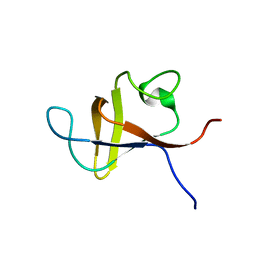 | | SOLUTION STRUCTURE AND LIGAND-BINDING SITE OF THE SH3 DOMAIN OF THE P85ALPHA SUBUNIT OF PHOSPHATIDYLINOSITOL 3-KINASE | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE P85-ALPHA SUBUNIT SH3 DOMAIN | | Authors: | Booker, G.W, Gout, I, Downing, A.K, Driscoll, P.C, Boyd, J, Waterfield, M.D, Campbell, I.D. | | Deposit date: | 1993-07-19 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and ligand-binding site of the SH3 domain of the p85 alpha subunit of phosphatidylinositol 3-kinase.
Cell(Cambridge,Mass.), 73, 1993
|
|
1PNM
 
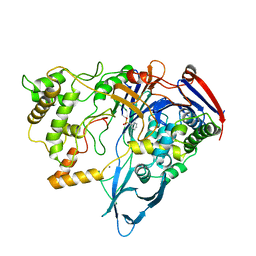 | |
1MUC
 
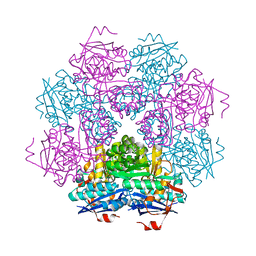 | | STRUCTURE OF MUCONATE LACTONIZING ENZYME AT 1.85 ANGSTROMS RESOLUTION | | Descriptor: | MANGANESE (II) ION, MUCONATE LACTONIZING ENZYME | | Authors: | Helin, S, Kahn, P.C, Guha, B.H.L, Mallows, D.J, Goldman, A. | | Deposit date: | 1995-09-20 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The refined X-ray structure of muconate lactonizing enzyme from Pseudomonas putida PRS2000 at 1.85 A resolution.
J.Mol.Biol., 254, 1995
|
|
1KN3
 
 | |
1NLI
 
 | | Complex of [E160A-E189A] trichosanthin and adenine | | Descriptor: | ADENINE, Ribosome-inactivating protein alpha-trichosanthin | | Authors: | Shaw, P.C, Wong, K.B, Chan, D.S.B, Williams, R.L. | | Deposit date: | 2003-01-07 | | Release date: | 2003-01-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural basis for the interaction of [E160A-E189A]-trichosanthin with adenine.
Toxicon, 41, 2003
|
|
1LR1
 
 | | Solution Structure of the Oligomerization Domain of the Bacterial Chromatin-Structuring Protein H-NS | | Descriptor: | dna-binding protein h-ns | | Authors: | Esposito, D, Petrovic, A, Harris, R, Ono, S, Eccleston, J, Mbabaali, A, Haq, I, Higgins, C.F, Hinton, J.C.D, Driscoll, P.C, Ladbury, J.E. | | Deposit date: | 2002-05-14 | | Release date: | 2003-01-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | H-NS Oligomerization Domain Structure Reveals the Mechanism for High Order
Self-association of the Intact Protein
J.Mol.Biol., 324, 2002
|
|
1PLB
 
 | | HIGH-RESOLUTION SOLUTION STRUCTURE OF REDUCED PARSLEY PLASTOCYANIN | | Descriptor: | COPPER (II) ION, PLASTOCYANIN | | Authors: | Bagby, S, Driscoll, P.C, Harvey, T.S, Hill, H.A.O. | | Deposit date: | 1994-05-20 | | Release date: | 1994-08-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of reduced parsley plastocyanin.
Biochemistry, 33, 1994
|
|
1PTS
 
 | |
1N4D
 
 | |
1MA0
 
 | | Ternary complex of Human glutathione-dependent formaldehyde dehydrogenase with NAD+ and dodecanoic acid | | Descriptor: | Glutathione-dependent formaldehyde dehydrogenase, LAURIC ACID, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Sanghani, P.C, Robinson, H, Bosron, W.F, Hurley, T.D. | | Deposit date: | 2002-07-30 | | Release date: | 2002-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Human glutathione-dependent formaldehyde dehydrogenase. Structures of apo, binary, and inhibitory ternary complexes.
Biochemistry, 41, 2002
|
|
1M7S
 
 | | Crystal Structure Analysis of Catalase CatF of Pseudomonas syringae | | Descriptor: | Catalase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Carpena, X, Soriano, M, Klotz, M.G, Duckworth, H.W, Donald, L.J, Melik-Adamyan, W, Fita, I, Loewen, P.C. | | Deposit date: | 2002-07-22 | | Release date: | 2002-08-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Clade 1 catalase, CatF of Pseudomonas syringae, at 1.8 A resolution
Proteins, 50, 2003
|
|
1RSZ
 
 | | Structure of human purine nucleoside phosphorylase in complex with DADMe-Immucillin-H and sulfate | | Descriptor: | 7-[[(3R,4R)-3-(hydroxymethyl)-4-oxidanyl-pyrrolidin-1-ium-1-yl]methyl]-3,5-dihydropyrrolo[3,2-d]pyrimidin-4-one, Purine nucleoside phosphorylase, SULFATE ION | | Authors: | Shi, W, Lewandowicz, A, Tyler, P.C, Furneaux, R.H, Almo, S.C, Schramm, V.L. | | Deposit date: | 2003-12-10 | | Release date: | 2005-02-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural comparison of human and malarial purine nucleoside phosphorylases
To be Published
|
|
1NG4
 
 | | Structure of ThiO (glycine oxidase) from Bacillus subtilis | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Glycine oxidase, HYDROGEN PEROXIDE, ... | | Authors: | Settembre, E.C, Dorrestein, P.C, Park, J, Augustine, A, Begley, T.P, Ealick, S.E. | | Deposit date: | 2002-12-16 | | Release date: | 2003-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Mechanistic Studies on ThiO, a Glycine Oxidase Essential for Thiamin Biosynthesis in Bacillus subtilis
Biochemistry, 42, 2003
|
|
1NG3
 
 | | Complex of ThiO (glycine oxidase) with acetyl-glycine | | Descriptor: | ACETYLAMINO-ACETIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Glycine oxidase, ... | | Authors: | Settembre, E.C, Dorrestein, P.C, Park, J, Augustine, A, Begley, T.P, Ealick, S.E. | | Deposit date: | 2002-12-16 | | Release date: | 2003-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Mechanistic Studies on ThiO, a Glycine Oxidase Essential for Thiamin Biosynthesis in Bacillus subtilis
Biochemistry, 42, 2003
|
|
1RHO
 
 | | STRUCTURE OF RHO GUANINE NUCLEOTIDE DISSOCIATION INHIBITOR | | Descriptor: | RHO GDP-DISSOCIATION INHIBITOR 1, SULFATE ION | | Authors: | Keep, N.H, Moody, P.C.E, Roberts, G.C.K. | | Deposit date: | 1996-10-12 | | Release date: | 1997-10-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A modulator of rho family G proteins, rhoGDI, binds these G proteins via an immunoglobulin-like domain and a flexible N-terminal arm.
Structure, 5, 1997
|
|
