5CC4
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS T62E/V66E at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Skerritt, L.A, Bell-Upp, P.C, Schlessman, J.L, Garcia-Moreno E, B. | | Deposit date: | 2015-07-01 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS T62E/V66E at cryogenic temperature
To be Published
|
|
1E3Y
 
 | |
5C5M
 
 | |
1DQP
 
 | | CRYSTAL STRUCTURE OF GIARDIA GUANINE PHOSPHORIBOSYLTRANSFERASE COMPLEXED WITH IMMUCILLING | | Descriptor: | 1,4-DIDEOXY-1,4-IMINO-1-(S)-(9-DEAZAGUANIN-9-YL)-D-RIBITOL, GUANINE PHOSPHORIBOSYLTRANSFERASE, ISOPROPYL ALCOHOL | | Authors: | Shi, W, Munagala, N.R, Wang, C.C, Li, C.M, Tyler, P.C, Furneaux, R.H, Grubmeyer, C, Schramm, V.L, Almo, S.C. | | Deposit date: | 2000-01-04 | | Release date: | 2000-07-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of Giardia lamblia guanine phosphoribosyltransferase at 1.75 A(,).
Biochemistry, 39, 2000
|
|
1DSL
 
 | | GAMMA B CRYSTALLIN C-TERMINAL DOMAIN | | Descriptor: | GAMMA B CRYSTALLIN | | Authors: | Norledge, B.V, Mayr, E.-M, Glockshuber, R, Bateman, O.A, Slingsby, C, Jaenicke, R, Driessen, H.P.C. | | Deposit date: | 1996-02-01 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The X-ray structures of two mutant crystallin domains shed light on the evolution of multi-domain proteins.
Nat.Struct.Biol., 3, 1996
|
|
5A0Q
 
 | |
1DQN
 
 | | CRYSTAL STRUCTURE OF GIARDIA GUANINE PHOSPHORIBOSYLTRANSFERASE COMPLEXED WITH A TRANSITION STATE ANALOGUE | | Descriptor: | GUANINE PHOSPHORIBOSYLTRANSFERASE, ISOPROPYL ALCOHOL, MAGNESIUM ION, ... | | Authors: | Shi, W, Munagala, N.R, Wang, C.C, Li, C.M, Tyler, P.C, Furneaux, R.H, Grubmeyer, C, Schramm, V.L, Almo, S.C. | | Deposit date: | 2000-01-04 | | Release date: | 2000-07-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of Giardia lamblia guanine phosphoribosyltransferase at 1.75 A(,).
Biochemistry, 39, 2000
|
|
2IA5
 
 | | T4 polynucleotide kinase/phosphatase with bound sulfate and magnesium. | | Descriptor: | ARSENIC, MAGNESIUM ION, Polynucleotide kinase, ... | | Authors: | Zhu, H, Smith, P.C, Wang, L.K, Lima, C.D, Shuman, S. | | Deposit date: | 2006-09-07 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-function analysis of the 3' phosphatase component of T4 polynucleotide kinase/phosphatase.
Virology, 366, 2007
|
|
244D
 
 | | THE HIGH-RESOLUTION CRYSTAL STRUCTURE OF A PARALLEL-STRANDED GUANINE TETRAPLEX | | Descriptor: | CALCIUM ION, DNA (5'-D(*TP*GP*GP*GP*GP*T)-3'), SODIUM ION | | Authors: | Laughlan, G, Murchie, A.I.H, Norman, D.G, Moore, M.H, Moody, P.C.E, Lilley, D.M.J, Luisi, B. | | Deposit date: | 1995-10-19 | | Release date: | 1996-02-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The high-resolution crystal structure of a parallel-stranded guanine tetraplex.
Science, 265, 1994
|
|
5CGK
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V66D/N100E at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Skerritt, L.A, Bell-Upp, P.C, Schlessman, J.L, Garcia-Moreno E, B. | | Deposit date: | 2015-07-09 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS V66D/N100E at cryogenic temperature
To be Published
|
|
4QOO
 
 | | Structure of Bacillus pumilus catalase with resorcinol bound. | | Descriptor: | CHLORIDE ION, Catalase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Loewen, P.C. | | Deposit date: | 2014-06-20 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Unprecedented access of phenolic substrates to the heme active site of a catalase: Substrate binding and peroxidase-like reactivity of Bacillus pumilus catalase monitored by X-ray crystallography and EPR spectroscopy.
Proteins, 83, 2015
|
|
4QOR
 
 | | Structure of Bacillus pumilus catalase with chlorophenol bound. | | Descriptor: | 2-CHLOROPHENOL, CHLORIDE ION, Catalase, ... | | Authors: | Loewen, P.C. | | Deposit date: | 2014-06-20 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Unprecedented access of phenolic substrates to the heme active site of a catalase: Substrate binding and peroxidase-like reactivity of Bacillus pumilus catalase monitored by X-ray crystallography and EPR spectroscopy.
Proteins, 83, 2015
|
|
4QON
 
 | | Structure of Bacillus pumilus catalase with catechol bound. | | Descriptor: | CATECHOL, CHLORIDE ION, Catalase, ... | | Authors: | Loewen, P.C. | | Deposit date: | 2014-06-20 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unprecedented access of phenolic substrates to the heme active site of a catalase: Substrate binding and peroxidase-like reactivity of Bacillus pumilus catalase monitored by X-ray crystallography and EPR spectroscopy.
Proteins, 83, 2015
|
|
4QOQ
 
 | | Structure of Bacillus pumilus catalase with guaiacol bound | | Descriptor: | CHLORIDE ION, Catalase, Guaiacol, ... | | Authors: | Loewen, P.C. | | Deposit date: | 2014-06-20 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Unprecedented access of phenolic substrates to the heme active site of a catalase: Substrate binding and peroxidase-like reactivity of Bacillus pumilus catalase monitored by X-ray crystallography and EPR spectroscopy.
Proteins, 83, 2015
|
|
6BCB
 
 | |
7XTV
 
 | | The structure of MHET-bound TfCut S130A | | Descriptor: | 4-(2-hydroxyethyloxycarbonyl)benzoic acid, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yang, Y, Jiang, P.C, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Complete bio-degradation of poly(butylene adipate-co-terephthalate) via engineered cutinases.
Nat Commun, 14, 2023
|
|
7XTU
 
 | | The structure of TfCut S130A | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Yang, Y, Jiang, P.C, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Complete bio-degradation of poly(butylene adipate-co-terephthalate) via engineered cutinases.
Nat Commun, 14, 2023
|
|
7XTW
 
 | | The structure of IsPETase in complex with MHET | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-hydroxyethyloxycarbonyl)benzoic acid, GLYCEROL, ... | | Authors: | Yang, Y, Jiang, P.C, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Complete bio-degradation of poly(butylene adipate-co-terephthalate) via engineered cutinases.
Nat Commun, 14, 2023
|
|
7XTR
 
 | | The apo structure of the engineered TfCut | | Descriptor: | GLYCEROL, LITHIUM ION, SULFATE ION, ... | | Authors: | Yang, Y, Jiang, P.C, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Complete bio-degradation of poly(butylene adipate-co-terephthalate) via engineered cutinases.
Nat Commun, 14, 2023
|
|
7XTS
 
 | | The apo structure of the engineered TfCut S130A | | Descriptor: | SODIUM ION, alpha/beta hydrolase | | Authors: | Yang, Y, Jiang, P.C, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Complete bio-degradation of poly(butylene adipate-co-terephthalate) via engineered cutinases.
Nat Commun, 14, 2023
|
|
7XTT
 
 | | The structure of engineered TfCut S130A in complex with MHET | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-hydroxyethyloxycarbonyl)benzoic acid, SODIUM ION, ... | | Authors: | Yang, Y, Jiang, P.C, Huang, J.W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Complete bio-degradation of poly(butylene adipate-co-terephthalate) via engineered cutinases.
Nat Commun, 14, 2023
|
|
2KXG
 
 | | The solution structure of the squash aspartic acid proteinase inhibitor (SQAPI) | | Descriptor: | Aspartic protease inhibitor | | Authors: | Headey, S.J, Macaskill, U.K, Wright, M, Claridge, J.K, Edwards, P.J.B, Farley, P.C, Christeller, J.T, Laing, W.A, Pascal, S.M. | | Deposit date: | 2010-05-05 | | Release date: | 2010-06-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the squash aspartic acid proteinase inhibitor (SQAPI) and mutational analysis of pepsin inhibition.
J.Biol.Chem., 285, 2010
|
|
1Y6Q
 
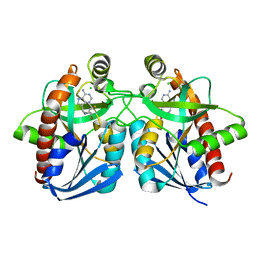 | | Cyrstal structure of MTA/AdoHcy nucleosidase complexed with MT-DADMe-ImmA | | Descriptor: | (3R,4S)-1-[(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)METHYL]-4-[(METHYLSULFANYL)METHYL]PYRROLIDIN-3-OL, CHLORIDE ION, MTA/SAH nucleosidase | | Authors: | Lee, J.E, Singh, V, Evans, G.B, Tyler, P.C, Furneaux, R.H, Cornell, K.A, Riscoe, M.K, Schramm, V.L, Howell, P.L. | | Deposit date: | 2004-12-06 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural rationale for the affinity of pico- and femtomolar transition state analogues of Escherichia coli 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase.
J.Biol.Chem., 280, 2005
|
|
5CR6
 
 | | Structure of pneumolysin at 1.98 A resolution | | Descriptor: | Pneumolysin | | Authors: | Marshall, J.E, Faraj, B.H.A, Gingras, A.R, Lonnen, R, Sheikh, M.A, El-Mezgueldi, M, Moody, P.C.E, Andrew, P.W, Wallis, R. | | Deposit date: | 2015-07-22 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The Crystal Structure of Pneumolysin at 2.0 angstrom Resolution Reveals the Molecular Packing of the Pre-pore Complex.
Sci Rep, 5, 2015
|
|
2A9E
 
 | |
