7JKU
 
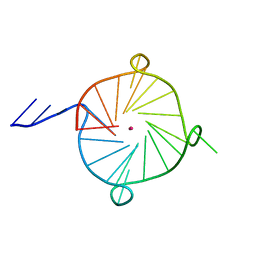 | | Crystal structure of a four-tetrad, parallel, and K+ stabilized Tetrahymena thermophila telomeric G-quadruplex | | Descriptor: | 26-mer DNA, POTASSIUM ION | | Authors: | Yatsunyk, L.A, McCarthy, S.E, Beseiso, D, Gallagher, E.P, Chen, E.V, Miao, J. | | Deposit date: | 2020-07-28 | | Release date: | 2021-07-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The first crystal structures of hybrid and parallel four-tetrad intramolecular G-quadruplexes.
Nucleic Acids Res., 50, 2022
|
|
4BNI
 
 | | Crystal structure of S. aureus FabI in complex with NADP and 2-(2- aminophenoxy)-5-hexylphenol | | Descriptor: | 2-(2-azanylphenoxy)-5-hexyl-phenol, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], GLUTAMIC ACID, ... | | Authors: | Schiebel, J, Chang, A, Bommineni, G.R, Tonge, P.J, Kisker, C. | | Deposit date: | 2013-05-15 | | Release date: | 2013-06-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rational Optimization of Drug-Target Residence Time: Insights from Inhibitor Binding to the S. Aureus Fabi Enzyme-Product Complex.
Biochemistry, 52, 2013
|
|
5VUW
 
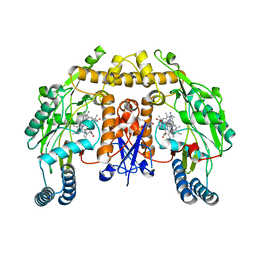 | | Structure of human neuronal nitric oxide synthase heme domain in complex with 7-(((3-(Dimethylamino)benzyl)amino)methyl)quinolin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[[[3-(dimethylamino)phenyl]methylamino]methyl]quinolin-2-amine, Nitric oxide synthase, ... | | Authors: | Huiying, L, Thomas, L.P. | | Deposit date: | 2017-05-19 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Hydrophilic, Potent, and Selective 7-Substituted 2-Aminoquinolines as Improved Human Neuronal Nitric Oxide Synthase Inhibitors.
J. Med. Chem., 60, 2017
|
|
3EY1
 
 | |
4TTT
 
 | |
4ITN
 
 | | Crystal structure of "compact P-loop" LpxK from Aquifex aeolicus in complex with chloride at 2.2 angstrom resolution | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Emptage, R.P, Pemble IV, C.W, York, J.D, Raetz, C.R.H, Zhou, P. | | Deposit date: | 2013-01-18 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1912 Å) | | Cite: | Mechanistic Characterization of the Tetraacyldisaccharide-1-phosphate 4'-Kinase LpxK Involved in Lipid A Biosynthesis.
Biochemistry, 52, 2013
|
|
4O21
 
 | | Product complex of metal-free PKAc, ATP-gamma-S and SP20. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Thio-phosphorylated peptide pSP20, cAMP-dependent protein kinase catalytic subunit alpha | | Authors: | Das, A, Kovalevsky, A.Y, Gerlits, O, Langan, P, Heller, W.T, Keshwani, M, Taylor, S. | | Deposit date: | 2013-12-16 | | Release date: | 2014-05-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Metal-Free cAMP-Dependent Protein Kinase Can Catalyze Phosphoryl Transfer.
Biochemistry, 53, 2014
|
|
5VUX
 
 | | Structure of human neuronal nitric oxide synthase heme domain in complex with 7-(((4-(Dimethylamino)benzyl)amino)methyl)quinolin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[({[4-(dimethylamino)phenyl]methyl}amino)methyl]quinolin-2-amine, Nitric oxide synthase, ... | | Authors: | Huiying, L, Thomas, L.P. | | Deposit date: | 2017-05-19 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Hydrophilic, Potent, and Selective 7-Substituted 2-Aminoquinolines as Improved Human Neuronal Nitric Oxide Synthase Inhibitors.
J. Med. Chem., 60, 2017
|
|
5W6P
 
 | | Crystal structure of Bacteriophage CBA120 tailspike protein 2 enzymatically active domain (TSP2dN, orf211) | | Descriptor: | 1,2-ETHANEDIOL, POTASSIUM ION, ZINC ION, ... | | Authors: | Plattner, M, Shneider, M.M, Leiman, P.G. | | Deposit date: | 2017-06-16 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.335 Å) | | Cite: | Structure and Function of the Branched Receptor-Binding Complex of Bacteriophage CBA120.
J.Mol.Biol., 431, 2019
|
|
3L29
 
 | | Crystal Structure of Zaire Ebola VP35 interferon inhibitory domain K319A/R322A mutant | | Descriptor: | CHLORIDE ION, Polymerase cofactor VP35 | | Authors: | Leung, D.W, Ramanan, P, Borek, D.M, Amarasinghe, G.K. | | Deposit date: | 2009-12-14 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mutations abrogating VP35 interaction with double-stranded RNA render ebola virus avirulent in guinea pigs.
J.Virol., 84, 2010
|
|
2V7F
 
 | | Structure of P. abyssi RPS19 protein | | Descriptor: | CHLORIDE ION, RPS19E SSU RIBOSOMAL PROTEIN S19E | | Authors: | Gregory, L.A, Aguissa-Toure, A.H, Pinaud, N, Legrand, P, Gleizes, P.E, Fribourg, S. | | Deposit date: | 2007-07-30 | | Release date: | 2007-09-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Molecular Basis of Diamond Blackfan Anemia: Structure and Function Analysis of Rps19.
Nucleic Acids Res., 35, 2007
|
|
4O1Y
 
 | | Crystal structure of Porcine Pancreatic Phospholipase A2 in complex with 1-Naphthaleneacetic acid | | Descriptor: | CALCIUM ION, NAPHTHALEN-1-YL-ACETIC ACID, Phospholipase A2, ... | | Authors: | Dileep, K.V, Remya, C, Tintu, I, Mandal, P.K, Karthe, P, Haridas, M, Sadasivan, C. | | Deposit date: | 2013-12-16 | | Release date: | 2014-01-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Porcine Pancreatic Phospholipase A2 in complex with 1-Naphthaleneacetic acid
To be published
|
|
2NXG
 
 | | Structural and mechanistic changes along an engineered path from metallo to non-metallo KDO8P synthase. | | Descriptor: | (2R,4R,5R,6R,7R)-2,4,5,6,7-PENTAHYDROXY-2,8-BIS(PHOSPHONOOXY)OCTANOIC ACID, 2-dehydro-3-deoxyphosphooctonate aldolase, ARABINOSE-5-PHOSPHATE, ... | | Authors: | Kona, F, Xu, X, Martin, P, Kuzmic, P, Gatti, D.L. | | Deposit date: | 2006-11-17 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Mechanistic Changes along an Engineered Path from Metallo to Nonmetallo 3-Deoxy-d-manno-octulosonate 8-Phosphate Synthases.
Biochemistry, 46, 2007
|
|
3WQK
 
 | | Crystal structure of Rv3378c with PO4 | | Descriptor: | Diterpene synthase, PHOSPHATE ION | | Authors: | Chan, H.C, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Zheng, Y, Bogue, S, Nakano, C, Hoshino, T, Zhang, L, Lv, P, Liu, W, Crick, D.C, Liang, P.H, Wang, A.H, Oldfield, E, Guo, R.T. | | Deposit date: | 2014-01-28 | | Release date: | 2014-02-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and inhibition of tuberculosinol synthase and decaprenyl diphosphate synthase from Mycobacterium tuberculosis.
J.Am.Chem.Soc., 136, 2014
|
|
3CWH
 
 | | D-xylose Isomerase in complex with linear product, per-deuterated xylulose | | Descriptor: | D-XYLULOSE, HYDROXIDE ION, MAGNESIUM ION, ... | | Authors: | Kovalevsky, A.Y, Langan, P, Glusker, J.P. | | Deposit date: | 2008-04-21 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | NEUTRON DIFFRACTION (2.2 Å) | | Cite: | Hydrogen location in stages of an enzyme-catalyzed reaction: time-of-flight neutron structure of D-xylose isomerase with bound D-xylulose
Biochemistry, 47, 2008
|
|
5W48
 
 | | Crystal Structure of Riboflavin Lyase (RcaE) | | Descriptor: | Riboflavin Lyase, SULFATE ION | | Authors: | Bhandari, D.M, Chakrabarty, Y, Zhao, B, Wood, J, Li, P, Begley, T.P. | | Deposit date: | 2017-06-09 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cannibalism Among the Flavins: a Novel C-N Bond Cleavage in Riboflavin Catabolism Mediated by Flavin-Generated Superoxide Radical
To be Published
|
|
1QF0
 
 | | THERMOLYSIN (E.C.3.4.24.27) COMPLEXED WITH (2-SULPHANYL-3-PHENYLPROPANOYL)-PHE-TYR. PARAMETERS FOR ZN-BIDENTATION OF MERCAPTOACYLDIPEPTIDES IN METALLOENDOPEPTIDASE | | Descriptor: | (2-SULFANYL-3-PHENYLPROPANOYL)-PHE-TYR, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Gaucher, J.-F, Selkti, M, Tiraboschi, G, Prange, T, Roques, B.P, Tomas, A, Fournie-Zaluski, M.C. | | Deposit date: | 1999-04-06 | | Release date: | 1999-12-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of alpha-mercaptoacyldipeptides in the thermolysin active site: structural parameters for a Zn monodentation or bidentation in metalloendopeptidases.
Biochemistry, 38, 1999
|
|
4O5Q
 
 | | Crystal Structure of the Alkylhydroperoxide Reductase AhpF from Escherichia coli | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alkyl hydroperoxide reductase subunit F, CADMIUM ION, ... | | Authors: | Dip, P.V, Kamariah, N, Manimekalai, M.S.S, Balakrishna, A.M, Gruber, G. | | Deposit date: | 2013-12-20 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure, mechanism and ensemble formation of the alkylhydroperoxide reductase subunits AhpC and AhpF from Escherichia coli
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5W67
 
 | | HLA-C*06:02 presenting VRSRR(ABA)LRL | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Mobbs, J.I, Vivian, J.P, Rossjohn, J. | | Deposit date: | 2017-06-16 | | Release date: | 2017-08-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The molecular basis for peptide repertoire selection in the human leucocyte antigen (HLA) C*06:02 molecule.
J. Biol. Chem., 292, 2017
|
|
5VV1
 
 | | Structure of human neuronal nitric oxide synthase heme domain in complex with 7-(((3-(4-Methoxypyridin-3-yl)propyl)amino)methyl)quinolin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-({[3-(4-methoxypyridin-3-yl)propyl]amino}methyl)quinolin-2-amine, Nitric oxide synthase, ... | | Authors: | Huiying, L, Thomas, L.P. | | Deposit date: | 2017-05-19 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Hydrophilic, Potent, and Selective 7-Substituted 2-Aminoquinolines as Improved Human Neuronal Nitric Oxide Synthase Inhibitors.
J. Med. Chem., 60, 2017
|
|
2O8D
 
 | | human MutSalpha (MSH2/MSH6) bound to ADP and a G dU mispair | | Descriptor: | 5'-D(*CP*CP*TP*AP*GP*CP*GP*(DU)P*GP*CP*GP*GP*TP*TP*C)-3', 5'-D(*GP*AP*AP*CP*CP*GP*CP*GP*CP*GP*CP*TP*AP*GP*G)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Warren, J.J, Pohlhaus, T.J, Changela, A, Modrich, P.L, Beese, L.S. | | Deposit date: | 2006-12-12 | | Release date: | 2007-06-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the Human MutSalpha DNA Lesion Recognition Complex.
Mol.Cell, 26, 2007
|
|
3LL8
 
 | |
4O5I
 
 | |
1ZGB
 
 | | Crystal Structure of Torpedo Californica Acetylcholinesterase in Complex With an (R)-Tacrine(10)-Hupyridone Inhibitor. | | Descriptor: | (5R)-5-{[10-(1,2,3,4-TETRAHYDROACRIDIN-9-YLAMINO)DECYL]AMINO}-5,6,7,8-TETRAHYDROQUINOLIN-2(1H)-ONE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase | | Authors: | Haviv, H, Wong, D.M, Greenblatt, H.M, Carlier, P.R, Pang, Y.P, Silman, I, Sussman, J.L, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2005-04-21 | | Release date: | 2005-08-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Packing Mediates Enantioselective Ligand Recognition at the Peripheral Site of Acetylcholinesterase
J.Am.Chem.Soc., 127, 2005
|
|
3D7V
 
 | | Crystal structure of Mcl-1 in complex with an Mcl-1 selective BH3 ligand | | Descriptor: | Bcl-2-like protein 11, Induced myeloid leukemia cell differentiation protein Mcl-1, ZINC ION | | Authors: | Lee, E.F, Czabotar, P.E, Colman, P.M, Fairlie, W.D. | | Deposit date: | 2008-05-22 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A novel BH3 ligand that selectively targets Mcl-1 reveals that apoptosis can proceed without Mcl-1 degradation.
J.Cell Biol., 180, 2008
|
|
