5HN2
 
 | |
7RHZ
 
 | |
7M58
 
 | |
7RLX
 
 | |
5HNJ
 
 | |
5HUM
 
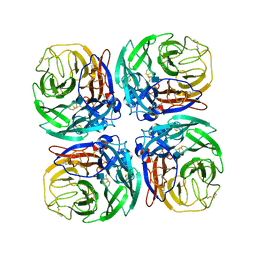 | | The crystal structure of neuraminidase from A/Sichuan/26221/2014 influenza virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, neuraminidase | | Authors: | Yang, H, Carney, P.J, Guo, Z, Chang, J.C, Stevens, J. | | Deposit date: | 2016-01-27 | | Release date: | 2016-04-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular Characterizations of Surface Proteins Hemagglutinin and Neuraminidase from Recent H5Nx Avian Influenza Viruses.
J.Virol., 90, 2016
|
|
3G2T
 
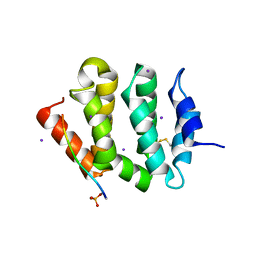 | | VHS Domain of human GGA1 complexed with SorLA C-terminal Phosphopeptide | | Descriptor: | ADP-ribosylation factor-binding protein GGA1, IODIDE ION, Phosphorylated C-terminal fragment of Sortilin-related receptor | | Authors: | Cramer, J.F, Behrens, M.A, Gustafsen, C, Oliveira, C.L.P, Pedersen, J.S, Madsen, P, Petersen, C.M, Thirup, S.S. | | Deposit date: | 2009-02-01 | | Release date: | 2009-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | GGA autoinhibition revisited
Traffic, 11, 2010
|
|
7MGA
 
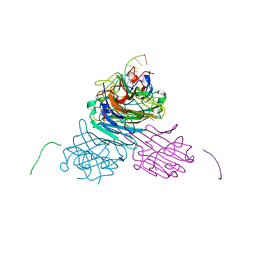 | | Concanavalin A bound to a DNA glycoconjugate, Man-AAATTT | | Descriptor: | 3-{[2-(2-hydroxyethoxy)ethyl]amino}-4-[(6-hydroxyhexyl)amino]cyclobut-3-ene-1,2-dione, CALCIUM ION, Concanavalin-A, ... | | Authors: | Partridge, B.E, Winegar, P.H, Mirkin, C.A. | | Deposit date: | 2021-04-12 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Redefining Protein Interfaces within Protein Single Crystals with DNA.
J.Am.Chem.Soc., 143, 2021
|
|
7MG5
 
 | | Concanavalin A bound to a DNA glycoconjugate, Man-ATAT | | Descriptor: | 3-{[2-(2-hydroxyethoxy)ethyl]amino}-4-[(6-hydroxyhexyl)amino]cyclobut-3-ene-1,2-dione, CALCIUM ION, Concanavalin-A, ... | | Authors: | Partridge, B.E, Winegar, P.H, Mirkin, C.A. | | Deposit date: | 2021-04-12 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Redefining Protein Interfaces within Protein Single Crystals with DNA.
J.Am.Chem.Soc., 143, 2021
|
|
5VYR
 
 | | Crystal structure of the WbkC formyl transferase from Brucella melitensis | | Descriptor: | (6R)-2-amino-6-methyl-5,6,7,8-tetrahydropteridin-4(3H)-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Riegert, A.S, Chantigian, D.P, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-05-26 | | Release date: | 2017-07-05 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical Characterization of WbkC, an N-Formyltransferase from Brucella melitensis.
Biochemistry, 56, 2017
|
|
6Z9X
 
 | | Human Class I Major Histocompatibility Complex, A02 allele, presenting LLS (t-butyl)Y FGTPT | | Descriptor: | Beta-2-microglobulin, LEU-LEU-SER-TUR-PHE-GLY-THR-PRO-THR, MHC class I antigen, ... | | Authors: | Rizkallah, P.J, Man, S, Redman, J.E. | | Deposit date: | 2020-06-04 | | Release date: | 2021-06-30 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Synthetic Peptides with Inadvertent Chemical Modifications Can Activate Potentially Autoreactive T Cells.
J Immunol., 207, 2021
|
|
6Z9Z
 
 | | Atomic resolution X-ray structure of the Uridine phosphorylase from Vibrio cholerae on crystals grown under microgravity | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Eistrikh-Heller, P.A, Rubinsky, S.V, Samygina, V.R, Gabdulkhakov, A.G, Lashkov, A.A. | | Deposit date: | 2020-06-04 | | Release date: | 2021-06-30 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Crystallization in Microgravity and the Atomic-Resolution Structure of Uridine Phosphorylase from Vibrio cholerae
Crystallography Reports, 66, 2021
|
|
6TPQ
 
 | | RNase M5 bound to 50S ribosome with precursor 5S rRNA | | Descriptor: | 50S ribosomal protein L10, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Oerum, S, Dendooven, T, Gilet, L, Catala, M, Degut, C, Trinquier, A, Barraud, P, Luisi, B, Condon, C, Tisne, C. | | Deposit date: | 2019-12-13 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structures of B. subtilis Maturation RNases Captured on 50S Ribosome with Pre-rRNAs.
Mol.Cell, 80, 2020
|
|
5VY2
 
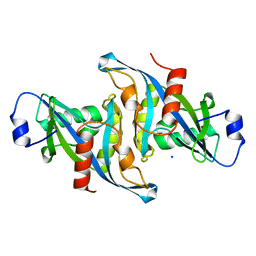 | | Crystal structure of the F36A mutant of HsNUDT16 | | Descriptor: | SODIUM ION, U8 snoRNA-decapping enzyme | | Authors: | Thirawatananond, P, Gabelli, S.B. | | Deposit date: | 2017-05-24 | | Release date: | 2018-11-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analyses of NudT16-ADP-ribose complexes direct rational design of mutants with improved processing of poly(ADP-ribosyl)ated proteins.
Sci Rep, 9, 2019
|
|
5HP4
 
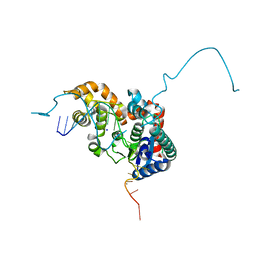 | | Crystal structure bacteriohage T5 D15 flap endonuclease (D155K) pseudo-enzyme-product complex with DNA and metal ions | | Descriptor: | CALCIUM ION, DNA (5'-D(*GP*AP*TP*CP*TP*AP*TP*AP*TP*GP*CP*CP*AP*TP*CP*GP*G)-3'), Exodeoxyribonuclease, ... | | Authors: | Almalki, F.A, Zhang, J, Sedelnikova, S.E, Rafferty, J.B, Sayers, J.R, Artymiuk, P.A. | | Deposit date: | 2016-01-20 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Direct observation of DNA threading in flap endonuclease complexes.
Nat.Struct.Mol.Biol., 23, 2016
|
|
8B70
 
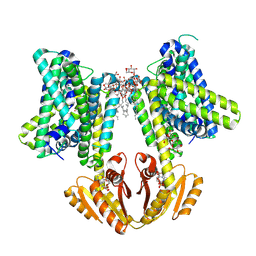 | | KimA from B. subtilis with nucleotide second-messenger c-di-AMP bound | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, DODECYL-BETA-D-MALTOSIDE, POTASSIUM ION, ... | | Authors: | Vonck, J, Wieferig, J.P. | | Deposit date: | 2022-09-28 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cyclic di-AMP traps proton-coupled K + transporters of the KUP family in an inward-occluded conformation.
Nat Commun, 14, 2023
|
|
6TQT
 
 | | The crystal structure of the MSP domain of human MOSPD2. | | Descriptor: | 1,2-ETHANEDIOL, Motile sperm domain-containing protein 2, PHOSPHATE ION | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Di Mattia, T, Wendling, C, Cavarelli, J, Tomasetto, C, Alpy, F. | | Deposit date: | 2019-12-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | FFAT motif phosphorylation controls formation and lipid transfer function of inter-organelle contacts.
Embo J., 39, 2020
|
|
7RA9
 
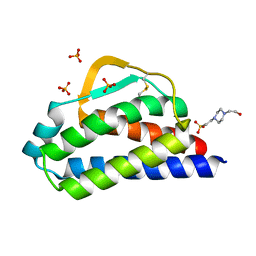 | | Designed StabIL-2 seq1 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Interleukin-2, PHOSPHATE ION | | Authors: | Jude, K.M, Chu, A.E, Huang, P.-S, Garcia, K.C. | | Deposit date: | 2021-06-30 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interleukin-2 superkines by computational design.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5W6F
 
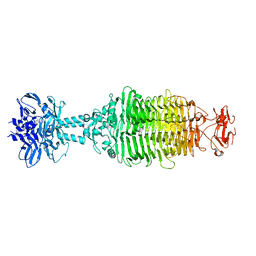 | |
8B71
 
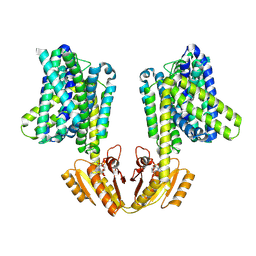 | | Upright KimA dimer with bound c-di-AMP from B. subtilis | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, Potassium transporter KimA | | Authors: | Vonck, J, Wieferig, J.P. | | Deposit date: | 2022-09-28 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cyclic di-AMP traps proton-coupled K + transporters of the KUP family in an inward-occluded conformation.
Nat Commun, 14, 2023
|
|
8BE1
 
 | | SARS-CoV-2 RBD in complex with a Fab fragment of a neutralising antibody mRBD2 | | Descriptor: | Antibody heavy chain, Antibody light chain, SULFATE ION, ... | | Authors: | Lulla, A, Brear, P, Fischer, K, Hollfelder, F, Hyvonen, M. | | Deposit date: | 2022-10-21 | | Release date: | 2023-01-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Microfluidics-enabled fluorescence-activated cell sorting of single pathogen-specific antibody secreting cells for the rapid discovery of monoclonal antibodies
Biorxiv, 2023
|
|
5W5F
 
 | | Cryo-EM structure of the T4 tail tube | | Descriptor: | Tail tube protein gp19 | | Authors: | Zheng, W, Wang, F, Taylor, N.M, Guerrero-Ferreira, R.C, Leiman, P.G, Egelman, E.H. | | Deposit date: | 2017-06-15 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Refined Cryo-EM Structure of the T4 Tail Tube: Exploring the Lowest Dose Limit.
Structure, 25, 2017
|
|
6ZH0
 
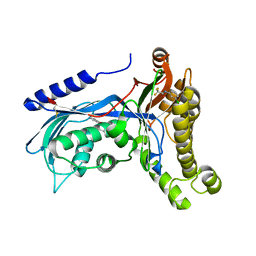 | | Structure of human galactokinase 1 bound with 2-(4-chlorophenyl)-N-(pyrimidin-2-yl)acetamide | | Descriptor: | 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, Galactokinase, N-(3-chlorophenyl)-2,2,2-trifluoroacetamide, ... | | Authors: | Mackinnon, S.R, Bezerra, G.A, Zhang, M, Foster, W, Krojer, T, Brandao-Neto, J, Douangamath, A, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, Lai, K, Yue, W.W. | | Deposit date: | 2020-06-20 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fragment Screening Reveals Starting Points for Rational Design of Galactokinase 1 Inhibitors to Treat Classic Galactosemia.
Acs Chem.Biol., 16, 2021
|
|
8RUF
 
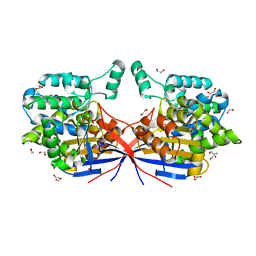 | | Crystal structure of Rhizobium etli L-asparaginase ReAV D187A mutant | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Pokrywka, K, Grzechowiak, M, Sliwiak, J, Worsztynowicz, P, Loch, J.I, Ruszkowski, M, Gilski, M, Jaskolski, M. | | Deposit date: | 2024-01-30 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Probing the active site of Class 3 L-asparaginase by mutagenesis. I. Tinkering with the zinc coordination site of ReAV.
Front Chem, 12, 2024
|
|
8RUA
 
 | | Crystal structure of Rhizobium etli L-asparaginase ReAV C135A mutant | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Pokrywka, K, Grzechowiak, M, Sliwiak, J, Worsztynowicz, P, Loch, J.I, Ruszkowski, M, Gilski, M, Jaskolski, M. | | Deposit date: | 2024-01-30 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Probing the active site of Class 3 L-asparaginase by mutagenesis. I. Tinkering with the zinc coordination site of ReAV.
Front Chem, 12, 2024
|
|
