7RX7
 
 | |
6WL1
 
 | | Cryo-EM of Form 1 related peptide filament, 36-31-3 | | Descriptor: | peptide 36-31-3 | | Authors: | Wang, F, Gnewou, O.M, Modlin, C, Egelman, E.H, Conticello, V.P. | | Deposit date: | 2020-04-17 | | Release date: | 2020-12-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural analysis of cross alpha-helical nanotubes provides insight into the designability of filamentous peptide nanomaterials.
Nat Commun, 12, 2021
|
|
7RX8
 
 | |
8F7T
 
 | | Glycan-Base ConC Env Trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 Env gp120, ... | | Authors: | Olia, A.S, Kwong, P.D. | | Deposit date: | 2022-11-20 | | Release date: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Soluble prefusion-closed HIV-envelope trimers with glycan-covered bases.
Iscience, 26, 2023
|
|
1AGO
 
 | |
6WL8
 
 | | Cryo-EM of Form 2 peptide filament | | Descriptor: | Form 2 peptide | | Authors: | Wang, F, Gnewou, O.M, Xu, C, Su, Z, Egelman, E.H, Conticello, V.P. | | Deposit date: | 2020-04-18 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural analysis of cross alpha-helical nanotubes provides insight into the designability of filamentous peptide nanomaterials.
Nat Commun, 12, 2021
|
|
8Q73
 
 | |
8D0I
 
 | | Human SARM1 bound to an NB-3 eADPR adduct | | Descriptor: | NAD(+) hydrolase SARM1, [[(2~{R},3~{S},4~{R},5~{R})-5-imidazo[2,1-f]purin-3-yl-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-[4-[(1~{S})-1-[methyl-[2,2,2-tris(fluoranyl)ethylcarbamoyl]amino]ethyl]pyridin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Bratkowski, M.A, Mathur, P. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Uncompetitive, adduct-forming SARM1 inhibitors are neuroprotective in preclinical models of nerve injury and disease.
Neuron, 110, 2022
|
|
7O76
 
 | |
7SSF
 
 | | Light harvesting phycobiliprotein HaPE560 from the cryptophyte Hemiselmis andersenii CCMP644 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DiCys-(15,16)-Dihydrobiliverdin, ... | | Authors: | Rathbone, H.W, Michie, K.A, Laos, A.L, Curmi, P.M.G. | | Deposit date: | 2021-11-10 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular dissection of the soluble photosynthetic antenna from the cryptophyte alga Hemiselmis andersenii
Commun Biol, 2023
|
|
6TJV
 
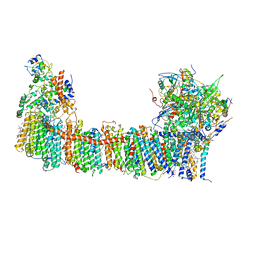 | | Structure of the NDH-1MS complex from Thermosynechococcus elongatus | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, BETA-CAROTENE, ... | | Authors: | Schuller, J.M, Saura, P, Thiemann, J, Schuller, S.K, Gamiz-Hernandez, A.P, Kurisu, G, Nowaczyk, M.M, Kaila, V.R.I. | | Deposit date: | 2019-11-27 | | Release date: | 2020-02-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Redox-coupled proton pumping drives carbon concentration in the photosynthetic complex I.
Nat Commun, 11, 2020
|
|
8D0C
 
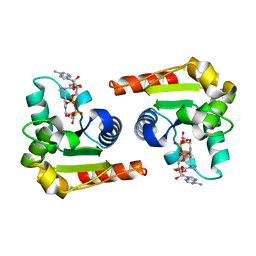 | | Human SARM1 TIR domain bound to NB-3-ADPR | | Descriptor: | NAD(+) hydrolase SARM1, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-[4-[(1~{S})-1-[methyl-[2,2,2-tris(fluoranyl)ethylcarbamoyl]amino]ethyl]pyridin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Bratkowski, M.A, Mathur, P. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Uncompetitive, adduct-forming SARM1 inhibitors are neuroprotective in preclinical models of nerve injury and disease.
Neuron, 110, 2022
|
|
8D0G
 
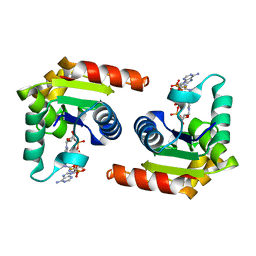 | | Human SARM1 TIR domain bound to NB-3-ADPRP | | Descriptor: | NAD(+) hydrolase SARM1, [[(2~{R},3~{R},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3-oxidanyl-4-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-[4-[(1~{S})-1-[methyl-[2,2,2-tris(fluoranyl)ethylcarbamoyl]amino]ethyl]pyridin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Bratkowski, M.A, Mathur, P. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Uncompetitive, adduct-forming SARM1 inhibitors are neuroprotective in preclinical models of nerve injury and disease.
Neuron, 110, 2022
|
|
8D0D
 
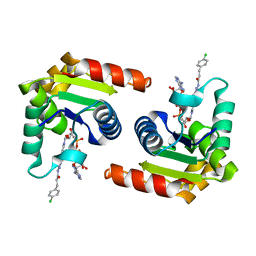 | | Human SARM1 TIR domain bound to an NB-7-ADPR adduct | | Descriptor: | NAD(+) hydrolase SARM1, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R})-5-[4-[3-[3-(4-chlorophenyl)propanoylamino]-4-methyl-1~{H}-pyrazol-5-yl]pyridin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Bratkowski, M.A, Mathur, P. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Uncompetitive, adduct-forming SARM1 inhibitors are neuroprotective in preclinical models of nerve injury and disease.
Neuron, 110, 2022
|
|
8D0H
 
 | | Human SARM1 TIR domain bound to NB-3-GDPR | | Descriptor: | NAD(+) hydrolase SARM1, [[(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-3~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-[4-[(1~{S})-1-[methyl-[2,2,2-tris(fluoranyl)ethylcarbamoyl]amino]ethyl]pyridin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Bratkowski, M.A, Mathur, P. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Uncompetitive, adduct-forming SARM1 inhibitors are neuroprotective in preclinical models of nerve injury and disease.
Neuron, 110, 2022
|
|
8D0F
 
 | | Human SARM1 TIR domain bound to NB-2-ADPR | | Descriptor: | NAD(+) hydrolase SARM1, [[(3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-[4-[[methyl-[2,2,2-tris(fluoranyl)ethylcarbamoyl]amino]methyl]pyridin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Bratkowski, M.A, Mathur, P. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Uncompetitive, adduct-forming SARM1 inhibitors are neuroprotective in preclinical models of nerve injury and disease.
Neuron, 110, 2022
|
|
8D0E
 
 | | Human SARM1 TIR domain bound to NB-7 | | Descriptor: | 3-(4-chlorophenyl)-N-[4-methyl-3-(pyridin-4-yl)-1H-pyrazol-5-yl]propanamide, NAD(+) hydrolase SARM1 | | Authors: | Bratkowski, M.A, Mathur, P. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Uncompetitive, adduct-forming SARM1 inhibitors are neuroprotective in preclinical models of nerve injury and disease.
Neuron, 110, 2022
|
|
8D0J
 
 | | Apo Human SARM1 TIR domain | | Descriptor: | NAD(+) hydrolase SARM1 | | Authors: | Bratkowski, M.A, Mathur, P. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Uncompetitive, adduct-forming SARM1 inhibitors are neuroprotective in preclinical models of nerve injury and disease.
Neuron, 110, 2022
|
|
6WKY
 
 | | Cryo-EM of Form 1 related peptide filament, 29-24-3 | | Descriptor: | peptide 29-24-3 | | Authors: | Wang, F, Gnewou, O.M, Egelman, E.H, Conticello, V.P. | | Deposit date: | 2020-04-17 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural analysis of cross alpha-helical nanotubes provides insight into the designability of filamentous peptide nanomaterials.
Nat Commun, 12, 2021
|
|
1AGZ
 
 | |
7OK5
 
 | | Crystal structure of mouse neurofascin 155 immunoglobulin domains | | Descriptor: | Neurofascin 155, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chataigner, L.M.P, Janssen, B.J.C. | | Deposit date: | 2021-05-17 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Structural insights into the contactin 1 - neurofascin 155 adhesion complex.
Nat Commun, 13, 2022
|
|
6YRV
 
 | | Crystal structure of FAP after illumination at 100K | | Descriptor: | CARBON DIOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, ... | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
6YRU
 
 | | Crystal structure of FAP in the dark at 100K | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, STEARIC ACID | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
6YS2
 
 | | Crystal structure of FAP R451A in the dark at 100K | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, STEARIC ACID | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
6YRX
 
 | | Low-dose crystal structure of FAP at room temperature | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, STEARIC ACID | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
