3IFX
 
 | | Crystal structure of the Spin-labeled KcsA mutant V48R1 | | Descriptor: | POTASSIUM ION, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate, TETRABUTYLAMMONIUM ION, ... | | Authors: | Cieslak, J.A, Focia, P.J, Gross, A. | | Deposit date: | 2009-07-26 | | Release date: | 2010-02-09 | | Last modified: | 2023-09-06 | | Method: | EPR (3.56 Å), X-RAY DIFFRACTION | | Cite: | Electron Spin-Echo Envelope Modulation (ESEEM) Reveals Water and Phosphate Interactions with the KcsA Potassium Channel
Biochemistry, 49, 2010
|
|
4HZF
 
 | | structure of the wild type Catabolite gene Activator Protein | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Catabolite gene activator, GLYCEROL, ... | | Authors: | Pohl, E, Townsend, P.D, Rodgers, T, Burnell, D, McLeish, T.C.B, Wilson, M.R, Cann, M. | | Deposit date: | 2012-11-15 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Modulation of global low-frequency motions underlies allosteric regulation: demonstration in CRP/FNR family transcription factors.
Plos Biol., 11, 2013
|
|
1ERY
 
 | | PHEROMONE ER-11, NMR | | Descriptor: | PHEROMONE ER-11 | | Authors: | Luginbuhl, P, Wu, J, Zerbe, O, Ortenzi, C, Luporini, P, Wuthrich, K. | | Deposit date: | 1996-03-22 | | Release date: | 1996-08-17 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structure of the pheromone Er-11 from the ciliated protozoan Euplotes raikovi.
Protein Sci., 5, 1996
|
|
1S3K
 
 | | Crystal Structure of a Humanized Fab (hu3S193) in Complex with the Lewis Y Tetrasaccharide | | Descriptor: | GLYCEROL, HU3S193 Fab fragment, heavy chain, ... | | Authors: | Ramsland, P.A, Farrugia, W, Bradford, T.M, Hogarth, P.M, Scott, A.M. | | Deposit date: | 2004-01-13 | | Release date: | 2004-07-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural convergence of antibody binding of carbohydrate determinants in lewis y tumor antigens
J.Mol.Biol., 340, 2004
|
|
1S44
 
 | | The structure and refinement of apocrustacyanin C2 to 1.6A resolution and the search for differences between this protein and the homologous apoproteins A1 and C1. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Crustacyanin A1 subunit, GLYCEROL, ... | | Authors: | Habash, J, Helliwell, J.R, Raftery, J, Cianci, M, Rizkallah, P.J, Chayen, N.E, Nneji, G.A, Zagalsky, P.F. | | Deposit date: | 2004-01-15 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure and refinement of apocrustacyanin C2 to 1.3 A resolution and the search for differences between this protein and the homologous apoproteins A1 and C1.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
2M7R
 
 | | Nmda receptor antagonist, conantokin bk-b, nmr, 20 structure | | Descriptor: | CONANTOKIN BK-B | | Authors: | Curtice, K.J, Platt, R.J, Skalicky, J.J, Horvath, M.P, Twede, V.D. | | Deposit date: | 2013-04-29 | | Release date: | 2014-02-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | From molecular phylogeny towards differentiating pharmacology for NMDA receptor subtypes.
Toxicon, 81C, 2014
|
|
1YSW
 
 | | Solution structure of the anti-apoptotic protein Bcl-2 complexed with an acyl-sulfonamide-based ligand | | Descriptor: | 3-NITRO-N-{4-[2-(2-PHENYLETHYL)-1,3-BENZOTHIAZOL-5-YL]BENZOYL}-4-{[2-(PHENYLSULFANYL)ETHYL]AMINO}BENZENESULFONAMIDE, Apoptosis regulator Bcl-2 | | Authors: | Oltersdorf, T, Elmore, S.W, Shoemaker, A.R, Armstrong, R.C, Augeri, D.J, Belli, B.A, Bruncko, M, Deckwerth, T.L, Dinges, J, Hajduk, P.J, Joseph, M.K, Kitada, S, Korsmeyer, S.J, Kunzer, A.R, Letai, A, Li, C, Mitten, M.J, Nettesheim, D.G, Ng, S, Nimmer, P.M, O'Connor, J.M, Oleksijew, A, Petros, A.M, Reed, J.C, Shen, W, Tahir, S.K, Thompson, C.B, Tomaselli, K.J, Wang, B, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H. | | Deposit date: | 2005-02-09 | | Release date: | 2005-06-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An inhibitor of Bcl-2 family proteins induces regression of solid tumours
Nature, 435, 2005
|
|
7AI4
 
 | |
2J8L
 
 | | FXI Apple 4 domain loop-out conformation | | Descriptor: | COAGULATION FACTOR XI | | Authors: | Samuel, D, Cheng, H, Riley, P.W, Canutescu, A.A, Bu, Z, Walsh, P.N, Roder, H. | | Deposit date: | 2006-10-25 | | Release date: | 2007-10-02 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the A4 Domain of Factor Xi Sheds Light on the Mechanism of Zymogen Activation.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
4E1K
 
 | | GlmU in complex with a Quinazoline Compound | | Descriptor: | Bifunctional protein GlmU, N-{4-[(7-hydroxy-6-methoxyquinazolin-4-yl)amino]phenyl}benzamide, SULFATE ION, ... | | Authors: | Larsen, N.A, Doig, P. | | Deposit date: | 2012-03-06 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An aminoquinazoline inhibitor of the essential bacterial cell wall synthetic enzyme GlmU has a unique non-protein-kinase-like binding mode.
Biochem.J., 446, 2012
|
|
1GYP
 
 | | CRYSTAL STRUCTURE OF GLYCOSOMAL GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE FROM LEISHMANIA MEXICANA: IMPLICATIONS FOR STRUCTURE-BASED DRUG DESIGN AND A NEW POSITION FOR THE INORGANIC PHOSPHATE BINDING SITE | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Kim, H, Feil, I.K, Verlinde, C.L.M.J, Petra, P.H, Hol, W.G.J. | | Deposit date: | 1995-08-01 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of glycosomal glyceraldehyde-3-phosphate dehydrogenase from Leishmania mexicana: implications for structure-based drug design and a new position for the inorganic phosphate binding site.
Biochemistry, 34, 1995
|
|
2BR8
 
 | | Crystal Structure of Acetylcholine-binding Protein (AChBP) from Aplysia californica in complex with an alpha-conotoxin PnIA variant | | Descriptor: | ALPHA-CONOTOXIN PNIA, SOLUBLE ACETYLCHOLINE RECEPTOR, SULFATE ION | | Authors: | Celie, P.H.N, Kasheverov, I.E, Mordvintsev, D.Y, Hogg, R.C, van Nierop, P, van Elk, R, van Rossum-Fikkert, S.E, Zhmak, M.N, Bertrand, D, Tsetlin, V, Sixma, T.K, Smit, A.B. | | Deposit date: | 2005-05-03 | | Release date: | 2005-06-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Nicotinic Acetylcholine Receptor Homolog Achbp in Complex with an Alpha-Conotoxin Pnia Variant
Nat.Struct.Mol.Biol., 12, 2005
|
|
1YNZ
 
 | | SH3 domain of yeast Pin3 | | Descriptor: | Pin3p | | Authors: | Kursula, P, Kursula, I, Zou, P, Lehmann, F, Song, Y.H, Wilmanns, M. | | Deposit date: | 2005-01-26 | | Release date: | 2006-05-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of the yeast SH3 domain proteome
To be Published
|
|
3RRW
 
 | | Crystal structure of the TL29 protein from Arabidopsis thaliana | | Descriptor: | CALCIUM ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Lundberg, E, Storm, P, Schroder, W.P, Funk, C. | | Deposit date: | 2011-05-01 | | Release date: | 2011-08-03 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the TL29 protein from Arabidopsis thaliana: An APX homolog without peroxidase activity.
J.Struct.Biol., 176, 2011
|
|
5O7Z
 
 | |
1S9O
 
 | | Solution structure of the nitrous acid induced DNA interstrand cross-linked dodecamer duplex CGCTAC(G)TAGCG with the cross-linked guanines denoted (G) | | Descriptor: | 5'-D(*CP*GP*CP*TP*AP*CP*GP*TP*AP*GP*CP*G)-3' | | Authors: | Edfeldt, N.B, Harwood, E.A, Sigurdsson, S.T, Hopkins, P.B, Reid, B.R. | | Deposit date: | 2004-02-05 | | Release date: | 2005-11-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a nitrous acid induced DNA interstrand cross-link.
Nucleic Acids Res., 32, 2004
|
|
1S8N
 
 | | Crystal structure of Rv1626 from Mycobacterium tuberculosis | | Descriptor: | AZIDE ION, putative antiterminator | | Authors: | Morth, J.P, Feng, V, Perry, L.J, Svergun, D.I, Tucker, P.A, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-02-03 | | Release date: | 2004-09-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.482 Å) | | Cite: | The Crystal and Solution Structure of a Putative Transcriptional Antiterminator from Mycobacterium tuberculosis.
Structure, 12, 2004
|
|
2MME
 
 | | Hybrid structure of the Shigella flexneri MxiH Type three secretion system needle | | Descriptor: | MxiH | | Authors: | Demers, J.P, Habenstein, B, Loquet, A, Vasa, S.K, Becker, S, Baker, D, Lange, A, Sgourakis, N.G. | | Deposit date: | 2014-03-14 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.7 Å), SOLID-STATE NMR | | Cite: | High-resolution structure of the Shigella type-III secretion needle by solid-state NMR and cryo-electron microscopy.
Nat Commun, 5, 2014
|
|
7KXS
 
 | | Computational design of constitutively active cGAS | | Descriptor: | Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Dowling, Q, Volkman, H.E, Gray, E.E, Ovchinnikov, S, Cambier, S, Bera, A.K, Bick, M, Kang, A, Stetson, D.B, King, N.P. | | Deposit date: | 2020-12-04 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Computational design of constitutively active cGAS.
Nat.Struct.Mol.Biol., 30, 2023
|
|
4MLT
 
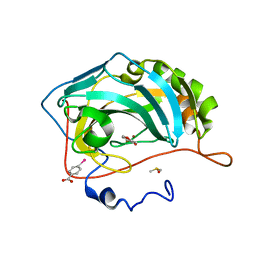 | |
2IVN
 
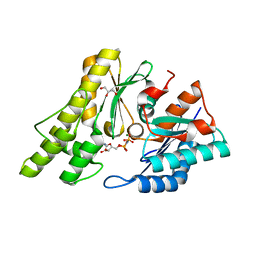 | | Structure of UP1 protein | | Descriptor: | GLYCEROL, MAGNESIUM ION, O-SIALOGLYCOPROTEIN ENDOPEPTIDASE, ... | | Authors: | Hecker, A, Leulliot, N, Graille, M, Dorlet, P, Quevillon-Cheruel, S, Ulryck, N, Van Tilbeurgh, H, Forterre, P. | | Deposit date: | 2006-06-14 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | An Archaeal Orthologue of the Universal Protein Kae1 is an Iron Metalloprotein which Exhibits Atypical DNA-Binding Properties and Apurinic-Endonuclease Activity in Vitro.
Nucleic Acids Res., 35, 2007
|
|
2XYM
 
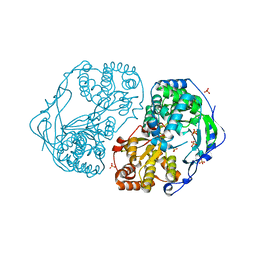 | | HCV-JFH1 NS5B T385A mutant | | Descriptor: | PHOSPHATE ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Simister, P.C, Caillet-Saguy, C, Bressanelli, S. | | Deposit date: | 2010-11-18 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.774 Å) | | Cite: | A Comprehensive Structure-Function Comparison of Hepatitis C Virus Strains Jfh1 and J6 Polymerases Reveals a Key Residue Stimulating Replication in Cell Culture Across Genotypes.
J.Virol., 85, 2011
|
|
1SB2
 
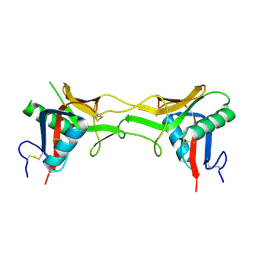 | | High resolution Structure determination of rhodocetin | | Descriptor: | Rhodocetin alpha subunit, Rhodocetin beta subunit | | Authors: | Paaventhan, P, Kong, C.G, Joseph, J.S, Chung, M.C.M, Kolatkar, P.R. | | Deposit date: | 2004-02-10 | | Release date: | 2005-02-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of rhodocetin reveals noncovalently bound heterodimer interface
Protein Sci., 14, 2005
|
|
5OHZ
 
 | |
4MNQ
 
 | | TCR-peptide specificity overrides affinity enhancing TCR-MHC interactions | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Rizkallah, P.J, Cole, D.K, Sewell, A.K, Jakobsen, B.K. | | Deposit date: | 2013-09-11 | | Release date: | 2013-11-13 | | Last modified: | 2017-08-02 | | Method: | X-RAY DIFFRACTION (2.742 Å) | | Cite: | T-cell receptor (TCR)-peptide specificity overrides affinity-enhancing TCR-major histocompatibility complex interactions.
J.Biol.Chem., 289, 2014
|
|
