3GEN
 
 | | The 1.6 A crystal structure of human bruton's tyrosine kinase bound to a pyrrolopyrimidine-containing compound | | Descriptor: | 4-Amino-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl-cyclopentane, Tyrosine-protein kinase BTK | | Authors: | Marcotte, D.J, Liu, Y.T, Arduini, R.M, Hession, C.A, Miatkowski, K, Wildes, C.P, Cullen, P.F, Hopkins, B, Mertsching, E, Jenkins, T.J, Romanowski, M.J, Baker, D.P, Silvian, L.F. | | Deposit date: | 2009-02-25 | | Release date: | 2010-01-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of human Bruton's tyrosine kinase in active and inactive conformations suggest a mechanism of activation for TEC family kinases.
Protein Sci., 19, 2010
|
|
1MYN
 
 | | SOLUTION STRUCTURE OF DROSOMYCIN, THE FIRST INDUCIBLE ANTIFUNGAL PROTEIN FROM INSECTS, NMR, 15 STRUCTURES | | Descriptor: | DROSOMYCIN | | Authors: | Landon, C, Sodano, P, Hetru, C, Hoffmann, J.A, Ptak, M. | | Deposit date: | 1996-12-26 | | Release date: | 1997-12-31 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of drosomycin, the first inducible antifungal protein from insects.
Protein Sci., 6, 1997
|
|
6FN1
 
 | | Zosuquidar and UIC2 Fab complex of human-mouse chimeric ABCB1 (ABCB1HM) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Human-mouse chimeric ABCB1 (ABCBHM), UIC2 Antigen Binding Fragment Heavy Chain, ... | | Authors: | Alam, A, Locher, K.P. | | Deposit date: | 2018-02-02 | | Release date: | 2018-02-21 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structure of a zosuquidar and UIC2-bound human-mouse chimeric ABCB1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5AG5
 
 | | CRYSTAL STRUCTURE OF LEISHMANIA MAJOR N-MYRISTOYLTRANSFERASE (NMT) WITH BOUND MYRISTOYL-COA AND A THIAZOLIDINONE LIGAND | | Descriptor: | (2R)-3-benzyl-2-(1H-indazol-5-yl)-1,3-thiazolidin-4-one, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, TETRADECANOYL-COA | | Authors: | Robinson, D.A, Spinks, D, Smith, V.C, Thompson, S, Smith, A, Torrie, L.S, McElroy, S.P, Brand, S, Brenk, R, Frearson, J.A, Read, K.D, Wyatt, P.G, Gilbert, I.H. | | Deposit date: | 2015-01-29 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of Small-Molecule Trypanosoma Brucei N-Myristoyltransferase Inhibitors: Discovery and Optimisation of a Novel Binding Mode.
Chemmedchem, 10, 2015
|
|
3ER8
 
 | | Crystal structure of the heterodimeric vaccinia virus mRNA polyadenylate polymerase complex with two fragments of RNA | | Descriptor: | Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase, Poly(A) polymerase catalytic subunit, RNA/DNA chimera 5'-D(CP*CP*)R(UP*UP*)D(C)-3', ... | | Authors: | Li, C, Li, H, Zhou, S, Poulos, T.L, Gershon, P.D. | | Deposit date: | 2008-10-01 | | Release date: | 2009-06-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Polymerase Translocation with Respect to Single-Stranded Nucleic Acid: Looping or Wrapping of Primer around a Poly(A) Polymerase
Structure, 17, 2009
|
|
4N25
 
 | | Crystal structure of Protein Arginine Deiminase 2 (250 uM Ca2+) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ... | | Authors: | Slade, D.J, Zhang, X, Fang, P, Dreyton, C.J, Zhang, Y, Gross, M.L, Guo, M, Coonrod, S.A, Thompson, P.R. | | Deposit date: | 2013-10-04 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.931 Å) | | Cite: | Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design.
Acs Chem.Biol., 10, 2015
|
|
5AG6
 
 | | CRYSTAL STRUCTURE OF LEISHMANIA MAJOR N-MYRISTOYLTRANSFERASE (NMT) WITH BOUND MYRISTOYL-COA AND A THIAZOLIDINONE LIGAND | | Descriptor: | (2R)-2-(4-hydroxy-3-methoxyphenyl)-3-(pyridin-2-ylmethyl)-1,3-thiazolidin-4-one, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, TETRADECANOYL-COA | | Authors: | Robinson, D.A, Spinks, D, Smith, V.C, Thompson, S, Smith, A, Torrie, L.S, McElroy, S.P, Brand, S, Brenk, R, Frearson, J.A, Read, K.D, Wyatt, P.G, Gilbert, I.H. | | Deposit date: | 2015-01-29 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of Small-Molecule Trypanosoma Brucei N-Myristoyltransferase Inhibitors: Discovery and Optimisation of a Novel Binding Mode.
Chemmedchem, 10, 2015
|
|
4N2N
 
 | | Crystal structure of Protein Arginine Deiminase 2 (E354A, 10 mM Ca2+) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Slade, D.J, Zhang, X, Fang, P, Dreyton, C.J, Zhang, Y, Gross, M.L, Guo, M, Coonrod, S.A, Thompson, P.R. | | Deposit date: | 2013-10-05 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design.
Acs Chem.Biol., 10, 2015
|
|
411D
 
 | | DUPLEX [5'-D(GCGTA+TACGC)]2 WITH INCORPORATED 2'-O-METHOXYETHYL RIBONUCLEOSIDE | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(T39)P*AP*CP*GP*C)-3') | | Authors: | Tereshko, V, Portmann, S, Tay, E.C, Martin, P, Natt, F, Altmann, K.H, Egli, M. | | Deposit date: | 1998-06-30 | | Release date: | 1998-07-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Correlating structure and stability of DNA duplexes with incorporated 2'-O-modified RNA analogues.
Biochemistry, 37, 1998
|
|
1N3C
 
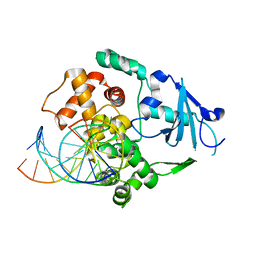 | | Structural and biochemical exploration of a critical amino acid in human 8-oxoguanine glycosylase | | Descriptor: | 8-oxoG-containing DNA, CALCIUM ION, DNA complement strand, ... | | Authors: | Norman, D.P, Chung, S.J, Verdine, G.L. | | Deposit date: | 2002-10-25 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and biochemical exploration of a critical amino acid in human 8-oxoguanine glycosylase
Biochemistry, 42, 2003
|
|
1JZW
 
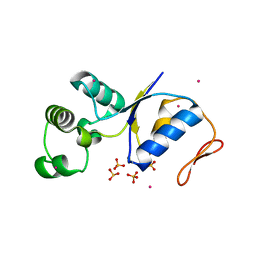 | | Arsenate Reductase + Sodium Arsenate From E. coli | | Descriptor: | ARSENATE REDUCTASE, CESIUM ION, SULFATE ION, ... | | Authors: | Martin, P, DeMel, S, Shi, J, Gladysheva, T, Gatti, D.L, Rosen, B.P, Edwards, B.F. | | Deposit date: | 2001-09-17 | | Release date: | 2001-11-28 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Insights into the structure, solvation, and mechanism of ArsC arsenate reductase, a novel arsenic detoxification enzyme.
Structure, 9, 2001
|
|
3IRN
 
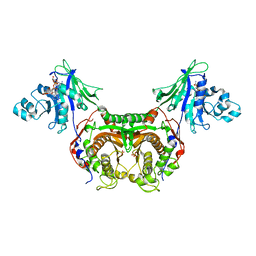 | | Trypanosoma cruzi Dihydrofolate Reductase-Thymidylate Synthase COMPLEXED WITH NADPH AND Cycloguanil | | Descriptor: | 1-(4-chlorophenyl)-6,6-dimethyl-1,6-dihydro-1,3,5-triazine-2,4-diamine, Bifunctional dihydrofolate reductase-thymidylate synthase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Chitnumsub, P, Yuvaniyama, J, Yuthavong, Y. | | Deposit date: | 2009-08-24 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of antifolate inhibition of Trypanosoma cruzi Dihydrofolate Reductase-Thymidylate Synthase
To be Published
|
|
1D40
 
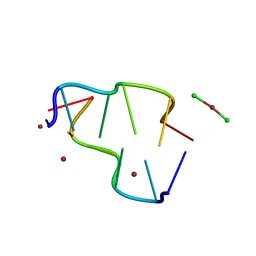 | | BASE SPECIFIC BINDING OF COPPER(II) TO Z-DNA: THE 1.3-ANGSTROMS SINGLE CRYSTAL STRUCTURE OF D(M5CGUAM5CG) IN THE PRESENCE OF CUCL2 | | Descriptor: | COPPER (II) CHLORIDE, COPPER (II) ION, DNA (5'-D(*(5CM)P*(CU)GP*UP*AP*(5CM)P*(CU)G)-3') | | Authors: | Geierstanger, B.H, Kagawa, T.F, Chen, S.-L, Quigley, G.J, Ho, P.S. | | Deposit date: | 1991-05-07 | | Release date: | 1992-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Base-specific binding of copper(II) to Z-DNA. The 1.3-A single crystal structure of d(m5CGUAm5CG) in the presence of CuCl2.
J.Biol.Chem., 266, 1991
|
|
3J07
 
 | | Model of a 24mer alphaB-crystallin multimer | | Descriptor: | Alpha-crystallin B chain | | Authors: | Jehle, S, Vollmar, B, Bardiaux, B, Dove, K.K, Rajagopal, P, Gonen, T, Oschkinat, H, Klevit, R.E. | | Deposit date: | 2011-04-27 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (20 Å), SOLID-STATE NMR, SOLUTION SCATTERING | | Cite: | N-terminal domain of {alpha}B-crystallin provides a conformational switch for multimerization and structural heterogeneity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2WYX
 
 | | Neutron structure of a class A Beta-lactamase Toho-1 E166A R274N R276N triple mutant | | Descriptor: | BETA-LACTAMSE TOHO-1 | | Authors: | Tomanicek, S.J, Blakeley, M.P, Cooper, J, Chen, Y, Afonine, P, Coates, L. | | Deposit date: | 2009-11-20 | | Release date: | 2010-01-12 | | Last modified: | 2024-05-08 | | Method: | NEUTRON DIFFRACTION (2.1 Å) | | Cite: | Neutron Diffraction Studies of a Class a Beta-Lactamase Toho-1 E166A R274N R276N Triple Mutant
J.Mol.Biol., 396, 2010
|
|
8DOZ
 
 | |
3ITC
 
 | | Crystal structure of Sco3058 with bound citrate and glycerol | | Descriptor: | CITRIC ACID, GLYCEROL, ZINC ION, ... | | Authors: | Nguyen, T.T, Cummings, J.A, Tsai, C.-L, Barondeau, D.P, Raushel, F.M. | | Deposit date: | 2009-08-28 | | Release date: | 2010-02-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure, mechanism, and substrate profile for Sco3058: the closest bacterial homologue to human renal dipeptidase
Biochemistry, 49, 2010
|
|
3QUL
 
 | | Crystal structures of the murine class I major histocompatibility complex H-2Db in complex with LCMV-derived gp33 altered peptide ligand (Y4S) | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Allerbring, E, Duru, A.D, Uchtenhagen, H, Madhurantakam, C, Grimm, S, Tomek, M.B, Mazumdar, P.A, Spetz, A, Friemann, R, Sandalova, T, Uhlin, M, Nygren, P, Achour, A. | | Deposit date: | 2011-02-24 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Unexpected T-cell recognition of an altered peptide ligand is driven by reversed thermodynamics.
Eur.J.Immunol., 42, 2012
|
|
3V46
 
 | | Crystal Structure of Yeast Cdc73 C-Terminal Domain | | Descriptor: | Cell division control protein 73 | | Authors: | Amrich, C.G, VanDemark, A.P. | | Deposit date: | 2011-12-14 | | Release date: | 2012-02-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | Cdc73 subunit of Paf1 complex contains C-terminal Ras-like domain that promotes association of Paf1 complex with chromatin.
J.Biol.Chem., 287, 2012
|
|
6FOK
 
 | | Copper transporter OprC | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, COPPER (II) ION, Putative copper transport outer membrane porin OprC | | Authors: | Bhamidimarri, S.P, van den Berg, B. | | Deposit date: | 2018-02-07 | | Release date: | 2019-03-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Acquisition of ionic copper by the bacterial outer membrane protein OprC through a novel binding site.
Plos Biol., 19, 2021
|
|
1XUW
 
 | | Structural rationalization of a large difference in RNA affinity despite a small difference in chemistry between two 2'-O-modified nucleic acid analogs | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(NMT)P*AP*CP*GP*C)-3') | | Authors: | Pattanayek, R, Sethaphong, L, Pan, C, Prhavc, M, Prakash, T.P, Manoharan, M, Egli, M. | | Deposit date: | 2004-10-26 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural rationalization of a large difference in RNA affinity despite a small difference in chemistry between two 2'-O-modified nucleic acid analogues.
J.Am.Chem.Soc., 126, 2004
|
|
4NB2
 
 | | Crystal Structure of FosB from Staphylococcus aureus at 1.89 Angstrom Resolution - Apo structure | | Descriptor: | Metallothiol transferase FosB, SULFATE ION | | Authors: | Cook, P.D, Thompson, M.K, Goodman, M.C, Jagessar, K, Harp, J, Keithly, M.E, Armstrong, R.N. | | Deposit date: | 2013-10-22 | | Release date: | 2014-02-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure and Function of the Genomically Encoded Fosfomycin Resistance Enzyme, FosB, from Staphylococcus aureus.
Biochemistry, 53, 2014
|
|
1JNB
 
 | | CONNECTOR PROTEIN FROM BACTERIOPHAGE PHI29 | | Descriptor: | UPPER COLLAR PROTEIN | | Authors: | Simpson, A.A, Leiman, P.G, Tao, Y, He, Y, Badasso, M.O, Jardine, P.J, Anderson, D.L, Rossmann, M.G. | | Deposit date: | 2001-07-23 | | Release date: | 2001-08-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure determination of the head-tail connector of bacteriophage phi29.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
5B8D
 
 | | Crystal structure of a low occupancy fragment candidate (N-(4-Methyl-1,3-thiazol-2-yl)propanamide) bound adjacent to the ubiquitin binding pocket of the HDAC6 zinc-finger domain | | Descriptor: | FORMIC ACID, Histone deacetylase 6, SODIUM ION, ... | | Authors: | Harding, R.J, Tempel, W, Collins, P, Pearce, N, Brandao-Neto, J, Douangamath, A, Ravichandran, M, Schapira, M, Bountra, C, Edwards, A.M, von Delft, F, Santhakumar, V, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-14 | | Release date: | 2016-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Small Molecule Antagonists of the Interaction between the Histone Deacetylase 6 Zinc-Finger Domain and Ubiquitin.
J. Med. Chem., 60, 2017
|
|
3UNQ
 
 | | Bovine trypsin variant X(triplePhe227) in complex with small molecule inhibitor | | Descriptor: | 1,2-ETHANEDIOL, BENZAMIDINE, CALCIUM ION, ... | | Authors: | Tziridis, A, Neumann, P, Kolenko, P, Stubbs, M.T. | | Deposit date: | 2011-11-16 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Correlating structure and ligand affinity in drug discovery: a cautionary tale involving second shell residues.
Biol.Chem., 395, 2014
|
|
