4BR0
 
 | | rat NTPDase2 in complex with Ca AMPNP | | Descriptor: | 5'-O-[(R)-hydroxy(phosphonoamino)phosphoryl]adenosine, CALCIUM ION, ECTONUCLEOSIDE TRIPHOSPHATE DIPHOSPHOHYDROLASE 2, ... | | Authors: | Zebisch, M, Schaefer, P, Lauble, P, Straeter, N. | | Deposit date: | 2013-06-03 | | Release date: | 2013-07-17 | | Last modified: | 2013-12-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystallographic Snapshots Along the Reaction Pathway of Nucleoside Triphosphate Diphosphohydrolases
Structure, 21, 2013
|
|
4BRC
 
 | | Legionella pneumophila NTPDase1 crystal form II, closed, Mg AMPNP complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-O-[(R)-hydroxy(phosphonoamino)phosphoryl]adenosine, CHLORIDE ION, ... | | Authors: | Zebisch, M, Schaefer, P, Lauble, P, Straeter, N. | | Deposit date: | 2013-06-04 | | Release date: | 2013-07-17 | | Last modified: | 2013-12-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystallographic Snapshots Along the Reaction Pathway of Nucleoside Triphosphate Diphosphohydrolases
Structure, 21, 2013
|
|
1PDT
 
 | | PD235, PNA-DNA DUPLEX, NMR, 8 STRUCTURES | | Descriptor: | DNA (5'-D(*GP*AP*CP*AP*TP*AP*GP*C)-3', PEPTIDE NUCLEIC ACID (COOH-P(*G*C*T*A*T*G*T*C)-NH2) | | Authors: | Eriksson, M, Nielsen, P.E. | | Deposit date: | 1996-03-28 | | Release date: | 1996-10-14 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a peptide nucleic acid-DNA duplex.
Nat.Struct.Biol., 3, 1996
|
|
4BRK
 
 | | Legionella pneumophila NTPDase1 N302Y variant crystal form III (closed) in complex with MG UMPPNP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-O-[(R)-hydroxy{[(S)-hydroxy(phosphonoamino)phosphoryl]oxy}phosphoryl]uridine, CHLORIDE ION, ... | | Authors: | Zebisch, M, Schaefer, P, Lauble, P, Straeter, N. | | Deposit date: | 2013-06-04 | | Release date: | 2013-07-17 | | Last modified: | 2013-12-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallographic Snapshots Along the Reaction Pathway of Nucleoside Triphosphate Diphosphohydrolases
Structure, 21, 2013
|
|
5TCY
 
 | | A complex of the synthetic siderophore analogue Fe(III)-5-LICAM with CeuE (H227L variant), a periplasmic protein from Campylobacter jejuni. | | Descriptor: | Enterochelin uptake periplasmic binding protein, FE (III) ION, N,N'-pentane-1,5-diylbis(2,3-dihydroxybenzamide) | | Authors: | Wilde, E.J, Blagova, E, Hughes, A, Raines, D.J, Moroz, O.V, Turkenburg, J.P, Duhme-Klair, A.-K, Wilson, K.S. | | Deposit date: | 2016-09-16 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Interactions of the periplasmic binding protein CeuE with Fe(III) n-LICAM(4-) siderophore analogues of varied linker length.
Sci Rep, 7, 2017
|
|
2Q66
 
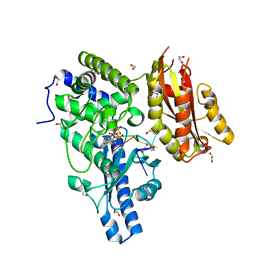 | | Structure of Yeast Poly(A) Polymerase with ATP and oligo(A) | | Descriptor: | 1,2-ETHANEDIOL, 5'-R(P*AP*AP*AP*AP*A)-3', ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Bohm, A, Balbo, P. | | Deposit date: | 2007-06-04 | | Release date: | 2007-08-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of poly(A) polymerase: structure of the enzyme-MgATP-RNA ternary complex and kinetic analysis.
Structure, 15, 2007
|
|
3FBV
 
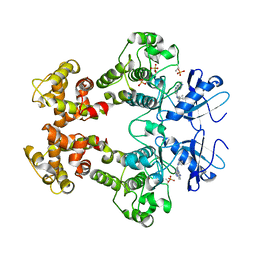 | | Crystal structure of the oligomer formed by the kinase-ribonuclease domain of Ire1 | | Descriptor: | N~2~-1H-benzimidazol-5-yl-N~4~-(3-cyclopropyl-1H-pyrazol-5-yl)pyrimidine-2,4-diamine, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Korennykh, A.V, Egea, P.F, Korostelev, A.A, Finer-Moore, J, Zhang, C, Shokat, K.M, Stroud, R.M, Walter, P. | | Deposit date: | 2008-11-19 | | Release date: | 2008-12-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The unfolded protein response signals through high-order assembly of Ire1.
Nature, 457, 2009
|
|
3C7N
 
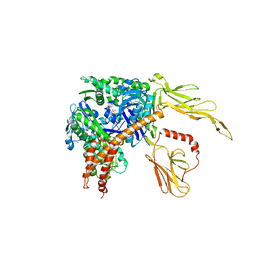 | | Structure of the Hsp110:Hsc70 Nucleotide Exchange Complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, CHLORIDE ION, ... | | Authors: | Schuermann, J.P, Jiang, J, Hart, P.J, Sousa, R. | | Deposit date: | 2008-02-07 | | Release date: | 2008-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.115 Å) | | Cite: | Structure of the Hsp110:Hsc70 nucleotide exchange machine
Mol.Cell, 31, 2008
|
|
6HFC
 
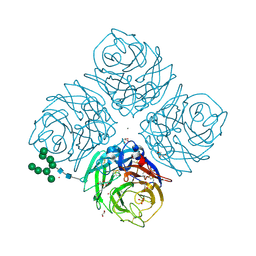 | | Influenza A Virus N9 Neuraminidase Native (Tern). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CARBON DIOXIDE, ... | | Authors: | Salinger, M.T, Hobbs, J.R, Murray, J.W, Laver, W.G, Kuhn, P, Garman, E.F. | | Deposit date: | 2018-08-21 | | Release date: | 2018-08-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | High Resolution Structures of Viral Neuraminidase with Drugs Bound in the Active Site. (In preparation)
To Be Published
|
|
3BNF
 
 | | W. succinogenes NrfA Sulfite Complex | | Descriptor: | ACETATE ION, CALCIUM ION, Cytochrome c-552, ... | | Authors: | Lukat, P, Einsle, O. | | Deposit date: | 2007-12-14 | | Release date: | 2008-02-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Binding and Reduction of Sulfite by Cytochrome c Nitrite Reductase
Biochemistry, 47, 2008
|
|
6HFY
 
 | | Influenza A virus N6 neuraminidase complex with DANA (Duck/England/56). | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Salinger, M.T, Hobbs, J.R, Murray, J.W, Laver, W.G, Kuhn, P, Garman, E.F. | | Deposit date: | 2018-08-22 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | High Resolution Structures of Viral Neuraminidase with Drugs Bound in the Active Site. (In preparation)
To Be Published
|
|
4BU1
 
 | | Crystal structure of Rad4 BRCT1,2 in complex with a Crb2 phosphopeptide | | Descriptor: | 1,2-ETHANEDIOL, DNA REPAIR PROTEIN RHP9, GLYCEROL, ... | | Authors: | Qu, M, Rappas, M, Wardlaw, C.P, Garcia, V, Carr, A.M, Oliver, A.W, Du, L.L, Pearl, L.H. | | Deposit date: | 2013-06-19 | | Release date: | 2013-10-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Phosphorylation-Dependent Assembly and Coordination of the DNA Damage Checkpoint Apparatus by Rad4(Topbp1.).
Mol.Cell, 51, 2013
|
|
2VFE
 
 | | Crystal structure of F96S mutant of Plasmodium falciparum triosephosphate isomerase with 3- phosphoglycerate bound at the dimer interface | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, GLYCEROL, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Gayathri, P, Banerjee, M, Vijayalakshmi, A, Balaram, H, Balaram, P, Murthy, M.R.N. | | Deposit date: | 2007-11-03 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical and Structural Characterization of Residue 96 Mutants of Plasmodium Falciparum Triosephosphate Isomerase: Active-Site Loop Conformation, Hydration and Identification of a Dimer-Interface Ligand-Binding Site.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
5HJO
 
 | | Murine endoplasmic reticulum alpha-glucosidase II with bound substrate analogue | | Descriptor: | 1,2-ETHANEDIOL, 2-deoxy-alpha-D-arabino-hexopyranose-(1-3)-D-glucal, ACETATE ION, ... | | Authors: | Caputo, A.T, Roversi, P, Alonzi, D.S, Kiappes, J.L, Zitzmann, N. | | Deposit date: | 2016-01-13 | | Release date: | 2016-07-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structures of mammalian ER alpha-glucosidase II capture the binding modes of broad-spectrum iminosugar antivirals.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1HJS
 
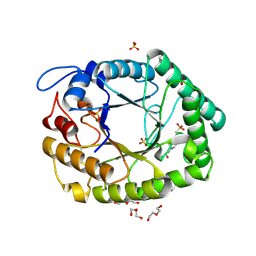 | | Structure of two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BETA-1,4-GALACTANASE, ... | | Authors: | Le Nours, J, Ryttersgaard, C, Lo Leggio, L, Ostergaard, P.R, Borchert, T.V, Christensen, L.L.H, Larsen, S. | | Deposit date: | 2003-02-27 | | Release date: | 2003-06-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure of Two Fungal Beta-1,4-Galactanases: Searching for the Basis for Temperature and Ph Optimum
Protein Sci., 12, 2003
|
|
1UPX
 
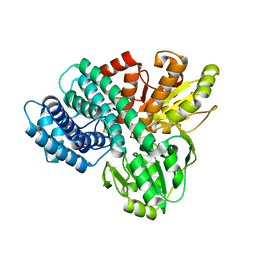 | | The crystal structure of the Hybrid Cluster Protein from Desulfovibrio desulfuricans containing molecules in the oxidized and reduced states. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FE4-S3 CLUSTER, HYDROXYLAMINE REDUCTASE, ... | | Authors: | Aragao, D, Macedo, S, Mitchell, E.P, Lindley, P.F. | | Deposit date: | 2003-10-14 | | Release date: | 2003-12-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structure of the Hybrid Cluster Protein (Hcp) from Desulfovibrio Desulfuricans Atcc 27774 Containing Molecules in the Oxidized and Reduced States
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1PL7
 
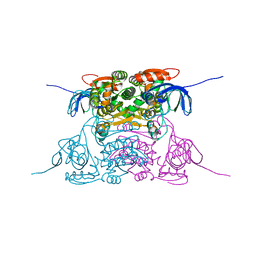 | | Human Sorbitol Dehydrogenase (apo) | | Descriptor: | Sorbitol dehydrogenase, ZINC ION | | Authors: | Pauly, T.A, Ekstrom, J.L, Beebe, D.A, Chrunyk, B, Cunningham, D, Griffor, M, Kamath, A, Lee, S.E, Madura, R, Mcguire, D, Subashi, T, Wasilko, D, Watts, P, Mylari, B.L, Oates, P.J, Adams, P.D, Rath, V.L. | | Deposit date: | 2003-06-07 | | Release date: | 2004-02-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray crystallographic and kinetic studies of human sorbitol dehydrogenase.
Structure, 11, 2003
|
|
6HG5
 
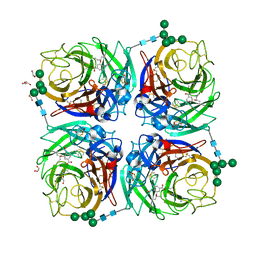 | | Influenza A virus N6 neuraminidase complex with Oseltamivir (Duck/England/56). | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Salinger, M.T, Hobbs, J.R, Murray, J.W, Laver, W.G, Kuhn, P, Garman, E.F. | | Deposit date: | 2018-08-22 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High Resolution Structures of Viral Neuraminidase with Drugs Bound in the Active Site.
To Be Published
|
|
4BR2
 
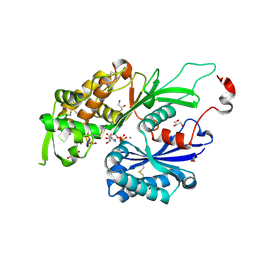 | | rat NTPDase2 in complex with Ca UMPPNP | | Descriptor: | 5'-O-[(R)-hydroxy{[(S)-hydroxy(phosphonoamino)phosphoryl]oxy}phosphoryl]uridine, CALCIUM ION, ECTONUCLEOSIDE TRIPHOSPHATE DIPHOSPHOHYDROLASE 2, ... | | Authors: | Zebisch, M, Schaefer, P, Lauble, P, Straeter, N. | | Deposit date: | 2013-06-03 | | Release date: | 2013-07-17 | | Last modified: | 2013-12-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic Snapshots Along the Reaction Pathway of Nucleoside Triphosphate Diphosphohydrolases
Structure, 21, 2013
|
|
4ZKO
 
 | |
4CBU
 
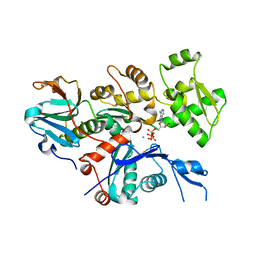 | | Crystal structure of Plasmodium falciparum actin I | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-1, CALCIUM ION, ... | | Authors: | Vahokoski, J, Bhargav, S.P, Desfosses, A, Andreadaki, M, Kumpula, E.P, Ignatev, A, Munico Martinez, S, Lepper, S, Frischknecht, F, Siden-Kiamos, I, Sachse, C, Kursula, I. | | Deposit date: | 2013-10-16 | | Release date: | 2014-04-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural Differences Explain Diverse Functions of Plasmodium Actins.
Plos Pathog., 10, 2014
|
|
5VYS
 
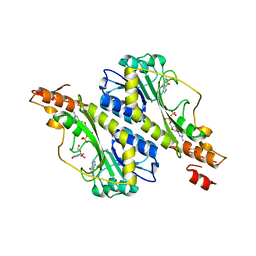 | | Crystal structure of the WbkC N-formyltransferase (C47S variant) from Brucella melitensis | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Riegert, A.S, Chantigian, D.P, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-05-26 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical Characterization of WbkC, an N-Formyltransferase from Brucella melitensis.
Biochemistry, 56, 2017
|
|
6MNZ
 
 | | Crystal structure of RibBX, a two domain 3,4-dihydroxy-2-butanone 4-phosphate synthase from A. baumannii. | | Descriptor: | 3,4-dihydroxy-2-butanone 4-phosphate synthase, CHLORIDE ION, SULFATE ION | | Authors: | Wang, J, Gonzalez-Gutierrez, G, Giedroc, D.P. | | Deposit date: | 2018-10-03 | | Release date: | 2019-04-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Multi-metal Restriction by Calprotectin Impacts De Novo Flavin Biosynthesis in Acinetobacter baumannii.
Cell Chem Biol, 26, 2019
|
|
5ORV
 
 | | Crystal structure of Aurora-A kinase in complex with an allosterically binding fragment | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Aurora kinase A, CHLORIDE ION, ... | | Authors: | McIntyre, P.J, Collins, P.M, von Delft, F, Bayliss, R. | | Deposit date: | 2017-08-16 | | Release date: | 2017-11-01 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Characterization of Three Druggable Hot-Spots in the Aurora-A/TPX2 Interaction Using Biochemical, Biophysical, and Fragment-Based Approaches.
ACS Chem. Biol., 12, 2017
|
|
5OS3
 
 | | Crystal structure of Aurora-A kinase in complex with an allosterically binding fragment | | Descriptor: | (1~{R})-1-(4-ethoxyphenyl)ethanamine, ADENOSINE-5'-DIPHOSPHATE, Aurora kinase A, ... | | Authors: | McIntyre, P.J, Collins, P.M, von Delft, F, Bayliss, R. | | Deposit date: | 2017-08-16 | | Release date: | 2017-11-01 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Characterization of Three Druggable Hot-Spots in the Aurora-A/TPX2 Interaction Using Biochemical, Biophysical, and Fragment-Based Approaches.
ACS Chem. Biol., 12, 2017
|
|
