1SEN
 
 | | Endoplasmic reticulum protein Rp19 O95881 | | Descriptor: | CHLORIDE ION, PLATINUM (II) ION, Thioredoxin-like protein p19 | | Authors: | Liu, Z.-J, Chen, L, Tempel, W, Shah, A, Lee, D, Dailey, T.A, Mayer, M.R, Rose, J.P, Richardson, D.C, Richardson, J.S, Dailey, H.A, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-02-17 | | Release date: | 2004-07-13 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.199 Å) | | Cite: | Endoplasmic reticulum protein Rp19
To be Published
|
|
4LBD
 
 | | LIGAND-BINDING DOMAIN OF THE HUMAN RETINOIC ACID RECEPTOR GAMMA BOUND TO THE SYNTHETIC AGONIST BMS961 | | Descriptor: | 3-FLUORO-4-[2-HYDROXY-2-(5,5,8,8-TETRAMETHYL-5,6,7,8,-TETRAHYDRO-NAPHTALEN-2-YL)-ACETYLAMINO]-BENZOIC ACID, RETINOIC ACID RECEPTOR GAMMA | | Authors: | Klaholz, B.P, Renaud, J.-P, Mitschler, A, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 1998-02-04 | | Release date: | 1999-03-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformational adaptation of agonists to the human nuclear receptor RAR gamma.
Nat.Struct.Biol., 5, 1998
|
|
5GJV
 
 | | Structure of the mammalian voltage-gated calcium channel Cav1.1 complex at near atomic resolution | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, J.P, Yan, Z, Li, Z.Q, Zhou, Q, Yan, N. | | Deposit date: | 2016-07-02 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of the voltage-gated calcium channel Cav1.1 at 3.6 angstrom resolution
Nature, 537, 2016
|
|
6JKI
 
 | | Crystal structure and catalytic mechanism of the essential m1G37 tRNA methyltransferase TrmD from Pseudomonas aeruginosa | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Jaroensuk, J, Liew, C.W, Atichartpongkul, S, Chionh, Y.H, Wong, Y.H, Zhong, W.H, McBee, M.E, Thongdee, N, Prestwich, E.G, DeMott, M.S, Mongkolsuk, S, Dedon, P.C, Lescar, J, Fuangthong, M. | | Deposit date: | 2019-03-01 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure and catalytic mechanism of the essential m1G37 tRNA methyltransferase TrmD fromPseudomonas aeruginosa.
Rna, 25, 2019
|
|
5CCM
 
 | | Crystal structure of SMYD3 with SAM and EPZ030456 | | Descriptor: | 6-chloranyl-2-oxidanylidene-N-[(1S,5R)-8-[4-[(phenylmethyl)amino]piperidin-1-yl]sulfonyl-8-azabicyclo[3.2.1]octan-3-yl]-1,3-dihydroindole-5-carboxamide, Histone-lysine N-methyltransferase SMYD3, S-ADENOSYLMETHIONINE, ... | | Authors: | Boriack-Sjodin, P.A. | | Deposit date: | 2015-07-02 | | Release date: | 2015-09-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Novel Oxindole Sulfonamides and Sulfamides: EPZ031686, the First Orally Bioavailable Small Molecule SMYD3 Inhibitor.
Acs Med.Chem.Lett., 7, 2016
|
|
8GNB
 
 | |
5JGN
 
 | | Spin-Labeled T4 Lysozyme Construct I9V1 | | Descriptor: | CHLORIDE ION, Endolysin, PHOSPHATE ION, ... | | Authors: | Balo, A.R, Feyrer, H, Ernst, O.P. | | Deposit date: | 2016-04-20 | | Release date: | 2017-02-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.534 Å) | | Cite: | Toward Precise Interpretation of DEER-Based Distance Distributions: Insights from Structural Characterization of V1 Spin-Labeled Side Chains.
Biochemistry, 55, 2016
|
|
3KJD
 
 | | Human poly(ADP-ribose) polymerase 2, catalytic fragment in complex with an inhibitor ABT-888 | | Descriptor: | (2R)-2-(7-carbamoyl-1H-benzimidazol-2-yl)-2-methylpyrrolidinium, GLYCEROL, Poly [ADP-ribose] polymerase 2 | | Authors: | Karlberg, T, Schutz, P, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Kotenyova, T, Kotzsch, A, Kraulis, P, Nielsen, T.K, Moche, M, Nordlund, P, Nyman, T, Persson, C, Roos, A.K, Siponen, M.I, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-11-03 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the catalytic domain of human PARP2 in complex with PARP inhibitor ABT-888.
Biochemistry, 49, 2010
|
|
4QOL
 
 | | Structure of Bacillus pumilus catalase | | Descriptor: | ACETATE ION, CHLORIDE ION, Catalase, ... | | Authors: | Loewen, P.C. | | Deposit date: | 2014-06-20 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Unprecedented access of phenolic substrates to the heme active site of a catalase: Substrate binding and peroxidase-like reactivity of Bacillus pumilus catalase monitored by X-ray crystallography and EPR spectroscopy.
Proteins, 83, 2015
|
|
5CFD
 
 | |
5JGR
 
 | | Spin-Labeled T4 Lysozyme Construct K43V1 | | Descriptor: | CHLORIDE ION, Endolysin, HEXANE-1,6-DIOL, ... | | Authors: | Balo, A.R, Feyrer, H, Ernst, O.P. | | Deposit date: | 2016-04-20 | | Release date: | 2017-02-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Toward Precise Interpretation of DEER-Based Distance Distributions: Insights from Structural Characterization of V1 Spin-Labeled Side Chains.
Biochemistry, 55, 2016
|
|
3KCL
 
 | |
5JGX
 
 | | Spin-Labeled T4 Lysozyme Construct V131V1 | | Descriptor: | CHLORIDE ION, Endolysin, PHOSPHATE ION, ... | | Authors: | Balo, A.R, Feyrer, H, Ernst, O.P. | | Deposit date: | 2016-04-20 | | Release date: | 2017-02-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.533 Å) | | Cite: | Toward Precise Interpretation of DEER-Based Distance Distributions: Insights from Structural Characterization of V1 Spin-Labeled Side Chains.
Biochemistry, 55, 2016
|
|
4QNN
 
 | |
3KKJ
 
 | | X-ray structure of P. syringae q888a4 Oxidoreductase at resolution 2.5A, Northeast Structural Genomics Consortium target PsR10 | | Descriptor: | Amine oxidase, flavin-containing, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Kuzin, A.P, Chen, Y, Forouhar, F, Vorobiev, S, Acton, T, Ma, L.C, Xiao, R, Montelione, G.T, Hunt, J.F, Tong, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-11-05 | | Release date: | 2009-11-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structure of P. syringae q888a4 Oxidoreductase at resolution 2.5A, Northeast Structural Genomics Consortium target PsR10
To be Published
|
|
5CDD
 
 | |
6SF7
 
 | | Atomic resolution structure of SplF protease from Staphylococcus aureus | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Golik, P, Stach, N, Karim, A, Dubin, G. | | Deposit date: | 2019-08-01 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Determinants of Substrate Specificity of SplF Protease from Staphylococcus aureus .
Int J Mol Sci, 22, 2021
|
|
8IYM
 
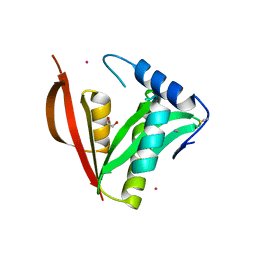 | | Crystal structure of a protein acetyltransferase, HP0935 | | Descriptor: | 1,2-ETHANEDIOL, N-acetyltransferase domain-containing protein, POTASSIUM ION, ... | | Authors: | Dadireddy, V, Mahanta, P, Kumar, A, Desirazu, R.N, Ramakumar, S. | | Deposit date: | 2023-04-05 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a protein acetyltransferase, HP0935
To be published
|
|
4QNK
 
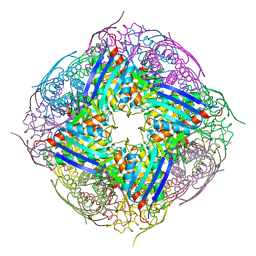 | | The structure of wt A. thaliana IGPD2 in complex with Mn2+ and phosphate | | Descriptor: | 1,2-ETHANEDIOL, Imidazoleglycerol-phosphate dehydratase 2, chloroplastic, ... | | Authors: | Bisson, C, Britton, K.L, Sedelnikova, S.E, Rodgers, H.F, Eadsforth, T.C, Viner, R, Hawkes, T.R, Baker, P.J, Rice, D.W. | | Deposit date: | 2014-06-18 | | Release date: | 2015-06-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structures Reveal that the Reaction Mechanism of Imidazoleglycerol-Phosphate Dehydratase Is Controlled by Switching Mn(II) Coordination.
Structure, 23, 2015
|
|
4QOQ
 
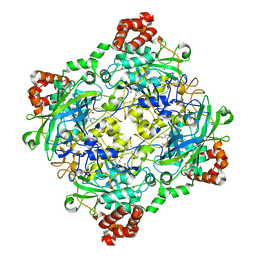 | | Structure of Bacillus pumilus catalase with guaiacol bound | | Descriptor: | CHLORIDE ION, Catalase, Guaiacol, ... | | Authors: | Loewen, P.C. | | Deposit date: | 2014-06-20 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Unprecedented access of phenolic substrates to the heme active site of a catalase: Substrate binding and peroxidase-like reactivity of Bacillus pumilus catalase monitored by X-ray crystallography and EPR spectroscopy.
Proteins, 83, 2015
|
|
1SM3
 
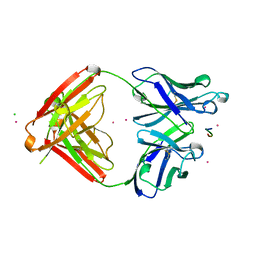 | | CRYSTAL STRUCTURE OF THE TUMOR SPECIFIC ANTIBODY SM3 COMPLEX WITH ITS PEPTIDE EPITOPE | | Descriptor: | CADMIUM ION, CHLORIDE ION, PEPTIDE EPITOPE, ... | | Authors: | Dokurno, P, Bates, P.A, Band, H.A, Stewart, L.M.D, Lally, J.M, Burchell, J.M, Taylor-Papadimitriou, J, Sternberg, M.J.E, Snary, D, Freemont, P.S. | | Deposit date: | 1997-12-23 | | Release date: | 1999-03-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure at 1.95 A resolution of the breast tumour-specific antibody SM3 complexed with its peptide epitope reveals novel hypervariable loop recognition.
J.Mol.Biol., 284, 1998
|
|
1WTT
 
 | |
1FQJ
 
 | | CRYSTAL STRUCTURE OF THE HETEROTRIMERIC COMPLEX OF THE RGS DOMAIN OF RGS9, THE GAMMA SUBUNIT OF PHOSPHODIESTERASE AND THE GT/I1 CHIMERA ALPHA SUBUNIT [(RGS9)-(PDEGAMMA)-(GT/I1ALPHA)-(GDP)-(ALF4-)-(MG2+)] | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(t) subunit alpha-1,Guanine nucleotide-binding protein G(i) subunit alpha-1,Guanine nucleotide-binding protein G(t) subunit alpha-1, MAGNESIUM ION, ... | | Authors: | Slep, K.C, Kercher, M.A, He, W, Cowan, C.W, Wensel, T.G, Sigler, P.B. | | Deposit date: | 2000-09-05 | | Release date: | 2001-02-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural determinants for regulation of phosphodiesterase by a G protein at 2.0 A.
Nature, 409, 2001
|
|
5MRB
 
 | | Crystal structure of human Mps1 (TTK) in complex with Cpd-5 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Dual specificity protein kinase TTK, ... | | Authors: | Hiruma, Y, Joosten, R.P, Perrakis, A. | | Deposit date: | 2016-12-22 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Understanding inhibitor resistance in Mps1 kinase through novel biophysical assays and structures.
J. Biol. Chem., 292, 2017
|
|
4ITL
 
 | | Crystal structure of LpxK from Aquifex aeolicus in complex with AMP-PCP at 2.1 angstrom resolution | | Descriptor: | GLYCEROL, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Tetraacyldisaccharide 4'-kinase | | Authors: | Emptage, R.P, Pemble IV, C.W, York, J.D, Raetz, C.R.H, Zhou, P. | | Deposit date: | 2013-01-18 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Mechanistic Characterization of the Tetraacyldisaccharide-1-phosphate 4'-Kinase LpxK Involved in Lipid A Biosynthesis.
Biochemistry, 52, 2013
|
|
