6T0D
 
 | | Crystal structure of YTHDC1 with fragment 27 (DHU_DC1_256) | | Descriptor: | SULFATE ION, YTHDC1, ~{N}-methyl-3-phenyl-1~{H}-pyrazole-5-carboxamide | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-02 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
5LH8
 
 | | Trypsin inhibitors for the treatment of pancreatitis - cpd 8 | | Descriptor: | (2~{S},4~{S})-1-[4-(aminomethyl)-3-methoxy-phenyl]carbonyl-4-(4-cyclopropyl-1,2,3-triazol-1-yl)-~{N}-[(1~{S},2~{R})-2-phenylcyclohexyl]pyrrolidine-2-carboxamide, CALCIUM ION, Cationic trypsin, ... | | Authors: | Schiering, N, D'Arcy, A, Skaanderup, P, Simic, O, Brandl, T, Woelcke, J. | | Deposit date: | 2016-07-08 | | Release date: | 2016-08-10 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Trypsin inhibitors for the treatment of pancreatitis.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5K12
 
 | | Cryo-EM structure of glutamate dehydrogenase at 1.8 A resolution | | Descriptor: | Glutamate dehydrogenase 1, mitochondrial | | Authors: | Merk, A, Bartesaghi, A, Banerjee, S, Falconieri, V, Rao, P, Earl, L, Milne, J, Subramaniam, S. | | Deposit date: | 2016-05-17 | | Release date: | 2016-06-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (1.8 Å) | | Cite: | Breaking Cryo-EM Resolution Barriers to Facilitate Drug Discovery.
Cell, 165, 2016
|
|
6T10
 
 | | Crystal structure of YTHDC1 with fragment 28 (DHU_DC1_176) | | Descriptor: | 5-iodanyl-~{N}-methyl-1~{H}-indazole-3-carboxamide, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-03 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
8CSA
 
 | |
2XED
 
 | | Nocardia farcinica maleate cis-trans isomerase C194S mutant with a covalently bound succinylcysteine intermediate | | Descriptor: | PUTATIVE MALEATE ISOMERASE, SUCCINIC ACID | | Authors: | Fisch, F, Martinez-Fleites, C, Baudendistel, N, Hauer, B, Turkenburg, J.P, Hart, S, Bruce, N.C, Grogan, G. | | Deposit date: | 2010-05-13 | | Release date: | 2010-08-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Covalent Succinylcysteine-Like Intermediate in the Enzyme-Catalyzed Transformation of Maleate to Fumarate by Maleate Isomerase.
J.Am.Chem.Soc., 132, 2010
|
|
5LIJ
 
 | | polyalanine chain built in bacteriophage phi812K1-420 cement protein density map | | Descriptor: | polyalanine chain built in bacteriophage phi812K1-420 cement protein density map | | Authors: | Novacek, J, Siborova, M, Benesik, M, Pantucek, R, Doskar, J, Plevka, P. | | Deposit date: | 2016-07-14 | | Release date: | 2017-07-26 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure and genome release of Twort-like Myoviridae phage with a double-layered baseplate.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
8CWP
 
 | | X-ray crystal structure of NTHi Protein D bound to a putative glycerol moiety | | Descriptor: | GLYCEROL, Glycerophosphoryl diester phosphodiesterase, SODIUM ION | | Authors: | Jones, S.P, Cook, K.H, Holmquist, M.L, Almekinder, L, DeLaney, A, Labbe, N, Perdue, J, Jackson, N, Charles, R, Pichichero, M, Kaur, R, Michel, L, Gleghorn, M.L. | | Deposit date: | 2022-05-19 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Vaccine target and carrier molecule nontypeable Haemophilus influenzae protein D dimerizes like the close Escherichia coli GlpQ homolog but unlike other known homolog dimers.
Proteins, 91, 2023
|
|
5YN5
 
 | | Crystal structure of MERS-CoV nsp10/nsp16 complex | | Descriptor: | ZINC ION, nsp10 protein, nsp16 protein | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
2XCW
 
 | |
3P28
 
 | |
6R2N
 
 | | Crystal structure of KlGlk1 glucokinase from Kluyveromyces lactis | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, Glucokinase-1 | | Authors: | Zak, K, Wator, E, Grudnik, P. | | Deposit date: | 2019-03-18 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.596 Å) | | Cite: | Crystal Structure of Kluyveromyces lactis Glucokinase ( Kl Glk1).
Int J Mol Sci, 20, 2019
|
|
2XKH
 
 | | Xe derivative of C.lacteus mini-Hb Leu86Ala mutant | | Descriptor: | NEURAL HEMOGLOBIN, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Pesce, A, Nardini, M, Dewilde, S, Capece, L, Marti, M.A, Congia, S, Salter, M.D, Blouin, G.C, Estrin, D.A, Ascenzi, P, Moens, L, Bolognesi, M, Olson, J.S. | | Deposit date: | 2010-07-08 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Ligand Migration in the Apolar Tunnel of Cerebratulus Lacteus Mini-Hemoglobin.
J.Biol.Chem., 286, 2011
|
|
6NNZ
 
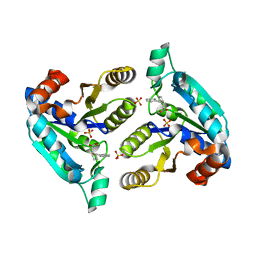 | | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with fragment analogue 4 | | Descriptor: | ATP-dependent dethiobiotin synthetase BioD, SULFATE ION, [(1R,2S)-2-(benzenecarbonyl)cyclopentyl]acetic acid, ... | | Authors: | Thompson, A.P, Polyak, S.W, Wegener, K.L, Bruning, J.B. | | Deposit date: | 2019-01-15 | | Release date: | 2020-01-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with fragment analogue 4
To Be Published
|
|
5YL4
 
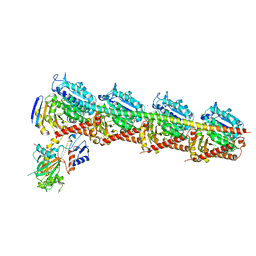 | | CRYSTAL STRUCTURE OF T2R-TTL-8WR COMPLEX | | Descriptor: | (3Z,6Z)-3-[(4-tert-butyl-1H-imidazol-5-yl)methylidene]-6-[[3-(phenylcarbonyl)phenyl]methylidene]piperazine-2,5-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Fu, Z.Y, Li, W.B, Chu, Y.Y, Hou, Y.W, Ji, C.P. | | Deposit date: | 2017-10-17 | | Release date: | 2017-11-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Design, synthesis and biological activity evaluation of plinabulin derivatives based on co-crystal structure
To Be Published
|
|
2XBS
 
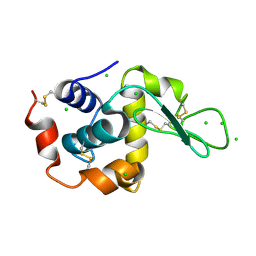 | | Raman crystallography of Hen White Egg Lysozyme - High X-ray dose (16 MGy) | | Descriptor: | CHLORIDE ION, LYSOZYME C | | Authors: | Carpentier, P, Royant, A, Weik, M, Bourgeois, D. | | Deposit date: | 2010-04-14 | | Release date: | 2010-11-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Raman Assisted Crystallography Reveals a Mechanism of X-Ray Induced Reversible Disulfide Radical Formation
Structure, 18, 2010
|
|
5K8S
 
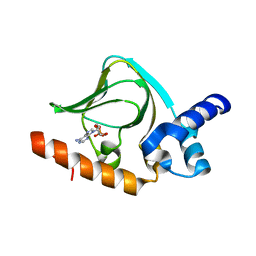 | | cAMP bound PfPKA-R (297-441) | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CAMP-dependent protein kinase regulatory subunit | | Authors: | Littler, D.R, Gilson, P.R, Crabb, B.S, Rossjohn, J.J. | | Deposit date: | 2016-05-31 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Disrupting the Allosteric Interaction between the Plasmodium falciparum cAMP-dependent Kinase and Its Regulatory Subunit.
J. Biol. Chem., 291, 2016
|
|
2XH3
 
 | | extracellular nuclease | | Descriptor: | DI(HYDROXYETHYL)ETHER, SPD1 NUCLEASE | | Authors: | Korczynska, J.E, Turkenburg, J.P, Taylor, E.J. | | Deposit date: | 2010-06-08 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | The Structural Characterization of a Prophage-Encoded Extracellular DNase from Streptococcus Pyogenes.
Nucleic Acids Res., 40, 2012
|
|
6NTJ
 
 | | Neutron/X-ray crystal structure of AAC-VIa bound to gentamicin C1A | | Descriptor: | (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDR OXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL, Aminoglycoside N(3)-acetyltransferase, MAGNESIUM ION | | Authors: | Cuneo, M.J, Kumar, P. | | Deposit date: | 2019-01-29 | | Release date: | 2019-09-25 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.9 Å), X-RAY DIFFRACTION | | Cite: | Low-Barrier and Canonical Hydrogen Bonds Modulate Activity and Specificity of a Catalytic Triad.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
3P8M
 
 | | Human dynein light chain (DYNLL2) in complex with an in vitro evolved peptide dimerized by leucine zipper | | Descriptor: | Dynein light chain 2, General control protein GCN4 | | Authors: | Rapali, P, Radnai, L, Suveges, D, Hetenyi, C, Harmat, V, Tolgyesi, F, Wahlgren, W.Y, Katona, G, Nyitray, L, Pal, G. | | Deposit date: | 2010-10-14 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Directed evolution reveals the binding motif preference of the LC8/DYNLL hub protein and predicts large numbers of novel binders in the human proteome.
Plos One, 6, 2011
|
|
2XD1
 
 | | ACTIVE SITE RESTRUCTURING REGULATES LIGAND RECOGNITION IN CLASS A PENICILLIN-BINDING PROTEINS | | Descriptor: | CEFOTAXIME, C3' cleaved, open, ... | | Authors: | Macheboeuf, P, Di Guilmi, A.M, Job, V, Vernet, T, Dideberg, O, Dessen, A. | | Deposit date: | 2010-04-28 | | Release date: | 2010-05-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Active Site Restructuring Regulates Ligand Recognition in Class a Penicillin-Binding Proteins.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2XJB
 
 | |
2XDG
 
 | | Crystal structure of the extracellular domain of human growth hormone releasing hormone receptor. | | Descriptor: | 1,2-ETHANEDIOL, GROWTH HORMONE-RELEASING HORMONE RECEPTOR, MAGNESIUM ION | | Authors: | Pike, A.C.W, Quigley, A, Barr, A.J, Burgess Brown, N, Shrestha, L, Yang, J, Chaikuad, A, Vollmar, M, Muniz, J.R.C, von Delft, F, Edwards, A, Arrowsmith, C.H, Weigelt, J, Bountra, C, Carpenter, E.P. | | Deposit date: | 2010-04-30 | | Release date: | 2010-06-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of the Extracellular Domain of Human Growth Hormone Releasing Hormone Receptor.
To be Published
|
|
2XCV
 
 | | Crystal structure of the D52N variant of cytosolic 5'-nucleotidase II in complex with inosine monophosphate and 2,3-bisphosphoglycerate | | Descriptor: | (2R)-2,3-diphosphoglyceric acid, CYTOSOLIC PURINE 5'-NUCLEOTIDASE, GLYCEROL, ... | | Authors: | Wallden, K, Nordlund, P. | | Deposit date: | 2010-04-26 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for the Allosteric Regulation and Substrate Recognition of Human Cytosolic 5'-Nucleotidase II
J.Mol.Biol., 408, 2011
|
|
6NP2
 
 | | AAC-VIa bound to Sisomicin | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(aminomethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, Aminoglycoside N(3)-acetyltransferase, MAGNESIUM ION | | Authors: | Kumar, P, Cuneo, M.J. | | Deposit date: | 2019-01-17 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Low-Barrier and Canonical Hydrogen Bonds Modulate Activity and Specificity of a Catalytic Triad.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
