2VWN
 
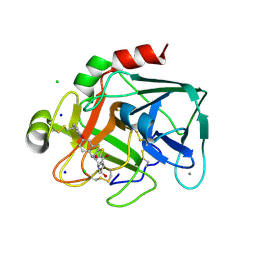 | | Aminopyrrolidine Factor Xa inhibitor | | Descriptor: | 5-Chloro-thiophene-2-carboxylic acid ((3S,4S)-1-{[2-fluoro-4-(2-oxo-2H-pyridin-1-yl)-phenylcarbamoyl]-methyl}-4-hydroxy-pyrrolidin-3-yl)-amide, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Groebke-Zbinden, K, Banner, D.W, Benz, J.M, Blasco, F, Decoret, G, Himber, J, Kuhn, B, Panday, N, Ricklin, F, Risch, P, Schlatter, D, Stahl, M, Unger, R, Haap, W. | | Deposit date: | 2008-06-26 | | Release date: | 2009-07-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Design of Novel Aminopyrrolidine Factor Xa Inhibitors from a Screening Hit.
Eur.J.Med.Chem., 44, 2009
|
|
5Y9G
 
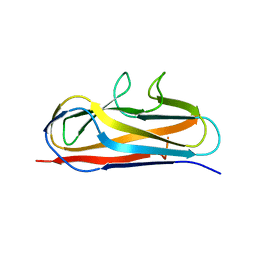 | | Crystal structure of Salmonella SafD adhesin | | Descriptor: | Pilin structural protein SafD,Pilus assembly protein | | Authors: | Zeng, L.H, Zhang, L, Wang, P.R, Meng, G. | | Deposit date: | 2017-08-25 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of host recognition and biofilm formation by Salmonella Saf pili
Elife, 6, 2017
|
|
5YBI
 
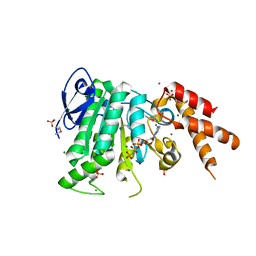 | | Structure of the bacterial pathogens ATPase with substrate AMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Probable ATP synthase SpaL/MxiB, ... | | Authors: | Mu, Z.X, Gao, X.P, Cui, S. | | Deposit date: | 2017-09-05 | | Release date: | 2018-06-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.268 Å) | | Cite: | Structural Insight Into Conformational Changes Induced by ATP Binding in a Type III Secretion-Associated ATPase FromShigella flexneri.
Front Microbiol, 9, 2018
|
|
8DKF
 
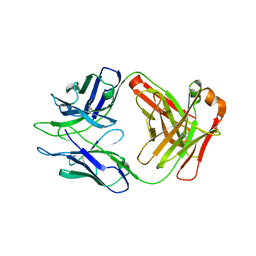 | | Antibody DH1030.1 Fab fragment | | Descriptor: | DH1030.1 Heavy chain, DH1030.1 Light chain | | Authors: | Gobeil, S, Acharya, P. | | Deposit date: | 2022-07-05 | | Release date: | 2023-07-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | DH1030.1 glycan reactive Ab
To Be Published
|
|
8J5J
 
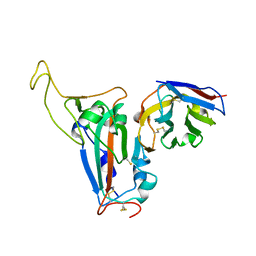 | | The crystal structure of bat coronavirus RsYN04 RBD bound to the antibody S43 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, nano antibody S43 | | Authors: | Zhao, R.C, Niu, S, Han, P, Qi, J.X, Gao, G.F, Wang, Q.H. | | Deposit date: | 2023-04-23 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cross-species recognition of bat coronavirus RsYN04 and cross-reaction of SARS-CoV-2 antibodies against the virus.
Zool.Res., 44, 2023
|
|
6SRH
 
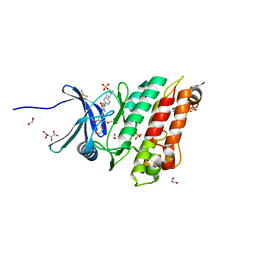 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound M4K2117 | | Descriptor: | 1,2-ETHANEDIOL, 4-methyl-3-[4-(1-methylpiperidin-4-yl)phenyl]-5-(3,4,5-trimethoxyphenyl)pyridine, Activin receptor type-1, ... | | Authors: | Adamson, R.J, Williams, E.P, Smil, D, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2019-09-05 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound M4K2117
To Be Published
|
|
8DP1
 
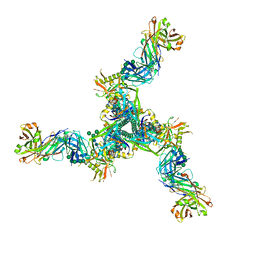 | | Cryo-EM structure of HIV-1 Env(BG505.T332N SOSIP) in complex with DH1030.1 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DH1030.1 Fab Heavy chain, ... | | Authors: | Gobeil, S, Acharya, P. | | Deposit date: | 2022-07-14 | | Release date: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Shared recognition mechanism for HIV-1 envelope-reactive V3 glycan broadly neutralizing B cell lineage maturation in humans and macaques
To Be Published
|
|
8DNQ
 
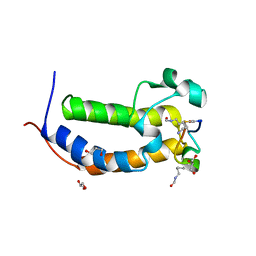 | | BRD2-BD1 in complex with cyclic peptide 2.2B | | Descriptor: | Bromodomain-containing protein 2, Cyclic peptide 2.2B, GLYCEROL | | Authors: | Patel, K, Franck, C, Mackay, J.P. | | Deposit date: | 2022-07-11 | | Release date: | 2023-07-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Discovery and characterization of cyclic peptides selective for the C-terminal bromodomains of BET family proteins.
Structure, 31, 2023
|
|
5YCT
 
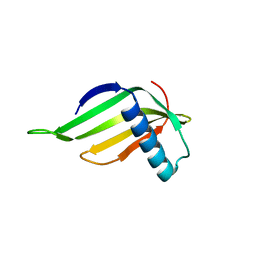 | | Engineered hairpin loop3 mutant monomer in Single-chain Monellin | | Descriptor: | Single chain Monellin | | Authors: | Surana, P, Nandwani, N, Udgaonkar, J.B, Gosavi, S, Das, R. | | Deposit date: | 2017-09-08 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | A five-residue motif for the design of domain swapping in proteins.
Nat Commun, 10, 2019
|
|
3OT1
 
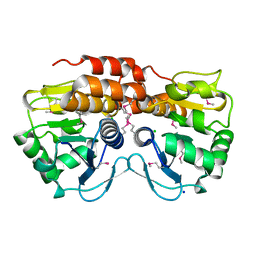 | | Crystal structure of VC2308 protein | | Descriptor: | 4-methyl-5(B-hydroxyethyl)-thiazole monophosphate biosynthesis enzyme, CHLORIDE ION, SODIUM ION | | Authors: | Niedzialkowska, E, Wawrzak, Z, Chruszcz, M, Porebski, P, Skarina, T, Huang, X, Grimshaw, S, Cymborowski, M, Savchenko, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-10 | | Release date: | 2010-09-22 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Crystal structure of VC2308 protein
To be Published
|
|
6SU7
 
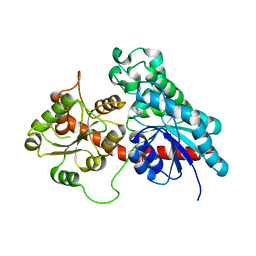 | | Complex between a UDP-glucosyltransferase from Polygonum tinctorium capable of glucosylating indoxyl and 3,4-Dichloroaniline | | Descriptor: | 3,4-Dichloroaniline, Glycosyltransferase | | Authors: | Fredslund, F, Teze, D, Svensson, B, Adams, P.D, Welner, D.H. | | Deposit date: | 2019-09-13 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | O-/N-/S-Specificity in Glycosyltransferase Catalysis: From Mechanistic Understanding to Engineering
Acs Catalysis, 11, 2021
|
|
5MOG
 
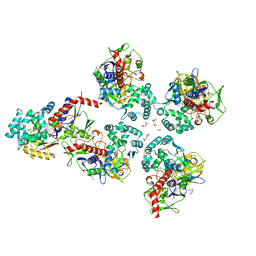 | | Oryza sativa phytoene desaturase inhibited by norflurazon | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Brausemann, A, Gemmecker, S, Koschmieder, J, Beyer, P, Einsle, O. | | Deposit date: | 2016-12-14 | | Release date: | 2017-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structure of Phytoene Desaturase Provides Insights into Herbicide Binding and Reaction Mechanisms Involved in Carotene Desaturation.
Structure, 25, 2017
|
|
8DLE
 
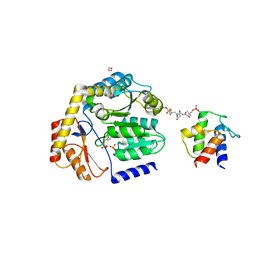 | | Crosslinked Crystal Structure of the 8-amino-7-oxonanoate synthase, BioF, and Benzene Sulfonyl Fluoride-crypto Acyl Carrier Protein, BSF-ACP | | Descriptor: | 1,2-ETHANEDIOL, 8-amino-7-oxononanoate synthase, Acyl carrier protein, ... | | Authors: | Chen, A, Davis, T.D, Louie, G.V, Bowman, M.E, Noel, J.P, Burkart, M.D. | | Deposit date: | 2022-07-07 | | Release date: | 2023-07-12 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Visualizing the Interface of Biotin and Fatty Acid Biosynthesis through SuFEx Probes.
J.Am.Chem.Soc., 146, 2024
|
|
2VZ5
 
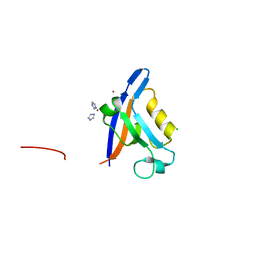 | | Structure of the PDZ domain of Tax1 (human T-cell leukemia virus type I) binding protein 3 | | Descriptor: | CHLORIDE ION, IMIDAZOLE, TAX1-BINDING PROTEIN 3, ... | | Authors: | Murray, J.W, Shafqat, N, Yue, W, Pilka, E, Johannsson, C, Salah, E, Cooper, C, Elkins, J.M, Pike, A.C, Roos, A, Filippakopoulos, P, von Delft, F, Wickstroem, M, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Oppermann, U. | | Deposit date: | 2008-07-30 | | Release date: | 2008-08-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.738 Å) | | Cite: | The Structure of the Pdz Domain of Tax1BP
To be Published
|
|
6P90
 
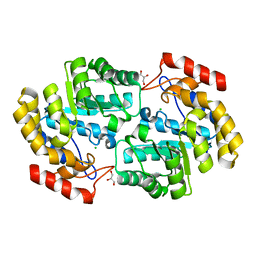 | | Crystal structure of PaDHDPS2-H56Q mutant | | Descriptor: | 4-hydroxy-tetrahydrodipicolinate synthase, CHLORIDE ION, GLYCEROL | | Authors: | Impey, R.E, Panjikar, S, Hall, C.J, Bock, L.J, Sutton, J.M, Perugini, M.A, Soares da Costa, T.P. | | Deposit date: | 2019-06-08 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of two dihydrodipicolinate synthase isoforms from Pseudomonas aeruginosa that differ in allosteric regulation.
Febs J., 287, 2020
|
|
8UC1
 
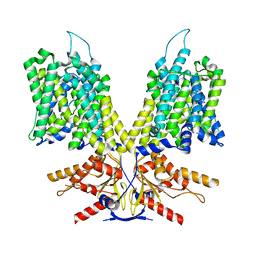 | |
5JD3
 
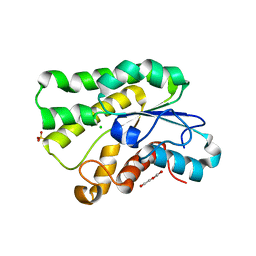 | | Crystal structure of LAE5, an alpha/beta hydrolase enzyme from the metagenome of Lake Arreo, Spain | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, LAE5, ... | | Authors: | Stogios, P.J, Xu, X, Nocek, B, Cui, H, Yim, V, Martinez-Martinez, M, Alcaide, M, Ferrer, M, Savchenko, A. | | Deposit date: | 2016-04-15 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | To be published
To Be Published
|
|
5MUT
 
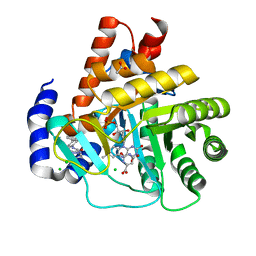 | | Crystal structure of potent human Dihydroorotate Dehydrogenase inhibitors based on hydroxylated azole scaffolds | | Descriptor: | 2-methyl-5-oxidanyl-~{N}-[2,3,5,6-tetrakis(fluoranyl)-4-phenyl-phenyl]-1,2,3-triazole-4-carboxamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Goyal, P, Andersson, M, Moritzer, A.C, Sainas, S, Pippione, A.C, Boschi, D, Al-Kadaraghi, S, Lolli, M, Friemann, R. | | Deposit date: | 2017-01-14 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Design, synthesis, biological evaluation and X-ray structural studies of potent human dihydroorotate dehydrogenase inhibitors based on hydroxylated azole scaffolds.
Eur J Med Chem, 129, 2017
|
|
8V7R
 
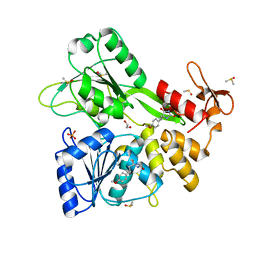 | | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z56772132 | | Descriptor: | (5R)-5-[2-(4-methoxyphenyl)ethyl]-5-methylimidazolidine-2,4-dione, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Godoy, A.S, Noske, G.D, Fairhead, M, Lithgo, R.M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Mesquita, N.C.M.R, Oliva, G, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-12-04 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z56772132
To Be Published
|
|
3ORQ
 
 | | Crystal Structure of N5-Carboxyaminoimidazole synthetase from Staphylococcus aureus complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Brugarolas, P, Duguid, E.M, Zhang, W, Poor, C.B, He, C. | | Deposit date: | 2010-09-07 | | Release date: | 2011-07-20 | | Last modified: | 2012-03-28 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural and biochemical characterization of N5-carboxyaminoimidazole ribonucleotide synthetase and N5-carboxyaminoimidazole ribonucleotide mutase from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
8DOW
 
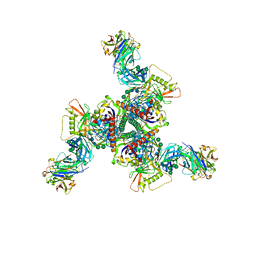 | | Cryo-EM structure of HIV-1 Env(CH848 10.17 DS.SOSIP_DT) in complex with DH1030.1 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DH1030.1 Fab Heavy chain, ... | | Authors: | Gobeil, S, Acharya, P. | | Deposit date: | 2022-07-14 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Shared recognition mechanism for HIV-1 envelope-reactive V3 glycan broadly neutralizing B cell lineage maturation in humans and macaques
To Be Published
|
|
2VOC
 
 | | THIOREDOXIN A ACTIVE SITE MUTANTS FORM MIXED DISULFIDE DIMERS THAT RESEMBLE ENZYME-SUBSTRATE REACTION INTERMEDIATE | | Descriptor: | DI(HYDROXYETHYL)ETHER, THIOREDOXIN | | Authors: | Kouwen, T.R.H.M, Andrell, J, Schrijver, R, Dubois, J.Y.F, Maher, M.J, Iwata, S, Carpenter, E.P, van Dijl, J.M. | | Deposit date: | 2008-02-13 | | Release date: | 2009-03-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Thioredoxin A active-site mutants form mixed disulfide dimers that resemble enzyme-substrate reaction intermediates.
J. Mol. Biol., 379, 2008
|
|
6SQW
 
 | | Mouse dCTPase in complex with 5-Me-dCMP | | Descriptor: | 5-METHYL-2'-DEOXY-CYTIDINE-5'-MONOPHOSPHATE, MAGNESIUM ION, dCTP pyrophosphatase 1 | | Authors: | Scaletti, E.R, Claesson, M, Helleday, H, Jemth, A.S, Stenmark, P. | | Deposit date: | 2019-09-04 | | Release date: | 2020-01-29 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The First Structure of an Active Mammalian dCTPase and its Complexes With Substrate Analogs and Products.
J.Mol.Biol., 432, 2020
|
|
5YNI
 
 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to SAM and m7GpppG | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, S-ADENOSYLMETHIONINE, ZINC ION, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Chen, Y, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
5YNQ
 
 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to SAH and m7GpppG | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-25 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.958 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
