2VBL
 
 | | Molecular basis of human XPC gene recognition and cleavage by engineered homing endonuclease heterodimers | | Descriptor: | 5'-D(*DA*DA*DA*DA*DG*DG*DC*DA*DG*DAP)-3', 5'-D(*DA*DG*DG*DA*DT*DC*DC*DT*DA*DAP)-3', 5'-D(*DT*DC*DT*DG*DC*DC*DT*DT*DT*DT*DT*DT *DGP*DAP)-3', ... | | Authors: | Redondo, P, Prieto, J, Munoz, I.G, Alibes, A, Stricher, F, Serrano, L, Arnould, S, Perez, C, Cabaniols, J.P, Duchateau, P, Paques, F, Blanco, F.J, Montoya, G. | | Deposit date: | 2007-09-14 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Basis of Xeroderma Pigmentosum Group C DNA Recognition by Engineered Meganucleases
Nature, 456, 2008
|
|
6OSS
 
 | | Crystal Structure of the Acyl-Carrier-Protein UDP-N-Acetylglucosamine O-Acyltransferase LpxA from Proteus mirabilis | | Descriptor: | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, PHOSPHITE ION, SULFATE ION | | Authors: | Kim, Y, Stogios, P, Skarina, T, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-05-02 | | Release date: | 2020-01-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal Structure of the Acyl-Carrier-Protein UDP-N-Acetylglucosamine O-Acyltransferase LpxA from Proteus mirabilis
To Be Published
|
|
1BR3
 
 | | CRYSTAL STRUCTURE OF AN 82-NUCLEOTIDE RNA-DNA COMPLEX FORMED BY THE 10-23 DNA ENZYME | | Descriptor: | DNA (10-23 DNA ENZYME), RNA (5'-R(*GP*GP*AP*CP*AP*GP*AP*UP*GP*GP*GP*AP*G)-3') | | Authors: | Nowakowski, J, Shim, P.J, Prasad, G.S, Stout, C.D, Joyce, G.F. | | Deposit date: | 1998-08-13 | | Release date: | 1999-02-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of an 82-nucleotide RNA-DNA complex formed by the 10-23 DNA enzyme.
Nat.Struct.Biol., 6, 1999
|
|
5LNX
 
 | |
6UW4
 
 | | Cryo-EM structure of human TRPV3 determined in lipid nanodisc | | Descriptor: | SODIUM ION, Transient receptor potential cation channel subfamily V member 3, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Deng, Z, Yuan, P. | | Deposit date: | 2019-11-04 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Gating of human TRPV3 in a lipid bilayer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6GYH
 
 | | Crystal structure of the light-driven proton pump Coccomyxa subellipsoidea Rhodopsin CsR | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHOLESTEROL, Family A G protein-coupled receptor-like protein, ... | | Authors: | Szczepek, M, Schmidt, A, Scheerer, P. | | Deposit date: | 2018-06-29 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design of a light-gated proton channel based on the crystal structure ofCoccomyxarhodopsin.
Sci.Signal., 12, 2019
|
|
7YMI
 
 | | PSII-Pcb Dimer of Acaryochloris Marina | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (6'R,11cis,11'cis,13cis,15cis)-4',5'-didehydro-5',6'-dihydro-beta,beta-carotene, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Shen, L.L, Gao, Y.Z, Wang, W.D, Zhang, X, Shen, J.R, Wang, P.Y, Han, G.Y. | | Deposit date: | 2022-07-28 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of a large photosystem II supercomplex from Acaryochloris marina.
To Be Published
|
|
7YMM
 
 | | PSII-Pcb Tetramer of Acaryochloris Marina | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (6'R,11cis,11'cis,13cis,15cis)-4',5'-didehydro-5',6'-dihydro-beta,beta-carotene, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Shen, L.L, Gao, Y.Z, Wang, W.D, Zhang, X, Shen, J.R, Wang, P.Y, Han, G.Y. | | Deposit date: | 2022-07-28 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of a large photosystem II supercomplex from Acaryochloris marina.
To Be Published
|
|
5ID0
 
 | | Cetuximab Fab in complex with aminoheptanoic acid-linked meditope | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cetuximab Fab heavy chain, Cetuximab Fab light chain, ... | | Authors: | Bzymek, K.P, Williams, J.C. | | Deposit date: | 2016-02-23 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Cyclization strategies of meditopes: affinity and diffraction studies of meditope-Fab complexes.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
7UVQ
 
 | | Pfs230 domain 1 bound by RUPA-97 and 15C5 Fabs | | Descriptor: | Fab Heavy Chain, Fab Kappa Light Chain, Gametocyte surface protein P230 | | Authors: | Ivanochko, D, Julien, J.P. | | Deposit date: | 2022-05-02 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Potent transmission-blocking monoclonal antibodies from naturally exposed individuals target a conserved epitope on Plasmodium falciparum Pfs230.
Immunity, 56, 2023
|
|
5LK3
 
 | |
5TCI
 
 | | Crystal structure of tryptophan synthase from M. tuberculosis - BRD4592-bound form | | Descriptor: | (2R,3S,4R)-3-(2'-fluoro[1,1'-biphenyl]-4-yl)-4-(hydroxymethyl)azetidine-2-carbonitrile, FORMIC ACID, MALONATE ION, ... | | Authors: | Michalska, K, Maltseva, N, Jedrzejczak, R, Wellington, S, Nag, P.P, Fisher, S.L, Schreiber, S.L, Hung, D.T, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-09-15 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A small-molecule allosteric inhibitor of Mycobacterium tuberculosis tryptophan synthase.
Nat. Chem. Biol., 13, 2017
|
|
6AU2
 
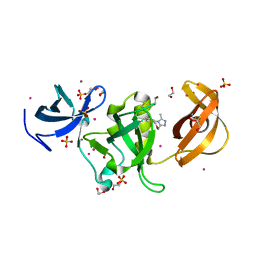 | | Crystal structure of SETDB1 Tudor domain with aryl triazole fragments | | Descriptor: | 1,2-ETHANEDIOL, 1-methyl-4H,6H-[1,2,4]triazolo[4,3-a][4,1]benzoxazepine, BETA-MERCAPTOETHANOL, ... | | Authors: | MADER, P, Mendoza-Sanchez, R, IQBAL, A, DONG, A, DOBROVETSKY, E, CORLESS, V.B, LIEW, S.K, TEMPEL, W, SMIL, D, DELA SENA, C.C, KENNEDY, S, DIAZ, D, HOLOWNIA, A, VEDADI, M, BROWN, P.J, SANTHAKUMAR, V, Bountra, C, Edwards, A.M, YUDIN, A.K, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-08-30 | | Release date: | 2017-10-11 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Identification and characterization of the first fragment hits for SETDB1 Tudor domain.
Bioorg.Med.Chem., 27, 2019
|
|
5LPU
 
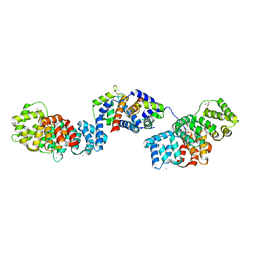 | | Crystal structure of Annexin A2 complexed with S100A4 | | Descriptor: | Annexin A2, CALCIUM ION, GLYCEROL, ... | | Authors: | Ecsedi, P, Gogl, G, Kiss, B, Nyitray, L. | | Deposit date: | 2016-08-15 | | Release date: | 2017-07-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Regulation of the Equilibrium between Closed and Open Conformations of Annexin A2 by N-Terminal Phosphorylation and S100A4-Binding.
Structure, 25, 2017
|
|
7AVM
 
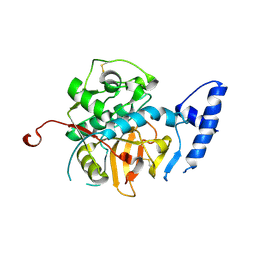 | | Crystal Structure of Pro-Rhodesain C150A | | Descriptor: | Cysteine protease, GLYCEROL | | Authors: | Johe, P, Jaenicke, E, Neuweiler, H, Schirmeister, T, Kersten, C, Hellmich, U. | | Deposit date: | 2020-11-05 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure, interdomain dynamics, and pH-dependent autoactivation of pro-rhodesain, the main lysosomal cysteine protease from African trypanosomes.
J.Biol.Chem., 296, 2021
|
|
5YG7
 
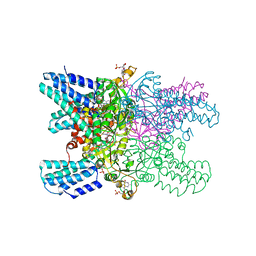 | | Crystal structure of ribose-1,5-bisphosphate isomerase mutant D204N from Pyrococcus horikoshii OT3 in complex with ribose-1,5-bisphosphate and GMP | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1,5-di-O-phosphono-alpha-D-ribofuranose, GUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Gogoi, P, Kanaujia, S.P. | | Deposit date: | 2017-09-22 | | Release date: | 2018-02-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A presumed homologue of the regulatory subunits of eIF2B functions as ribose-1,5-bisphosphate isomerase in Pyrococcus horikoshii OT3.
Sci Rep, 8, 2018
|
|
6UTQ
 
 | | LarE, a sulfur transferase involved in synthesis of the cofactor for lactate racemase in complex with cadmium | | Descriptor: | ATP-dependent sacrificial sulfur transferase LarE, CADMIUM ION, PHOSPHATE ION, ... | | Authors: | Fellner, M, Huizenga, K, Hausinger, R.P, Hu, J. | | Deposit date: | 2019-10-29 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystallographic characterization of a tri-Asp metal-binding site at the three-fold symmetry axis of LarE.
Sci Rep, 10, 2020
|
|
4I5C
 
 | | The Jak1 kinase domain in complex with inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 3-oxo-3-[(3R)-3-(pyrrolo[2,3-b][1,2,3]triazolo[4,5-d]pyridin-1(6H)-yl)piperidin-1-yl]propanenitrile, Tyrosine-protein kinase JAK1 | | Authors: | Fong, R, Lupardus, P.J. | | Deposit date: | 2012-11-28 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel triazolo-pyrrolopyridines as inhibitors of Janus kinase 1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3IIR
 
 | | Crystal Structure of Miraculin like protein from seeds of Murraya koenigii | | Descriptor: | Trypsin inhibitor | | Authors: | Gahloth, D, Selvakumar, P, Shee, C, Kumar, P, Sharma, A.K. | | Deposit date: | 2009-08-03 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cloning, sequence analysis and crystal structure determination of a miraculin-like protein from Murraya koenigii
Arch.Biochem.Biophys., 494, 2010
|
|
4XJQ
 
 | | The catalytic mechanism of human parainfluenza virus type 3 haemagglutinin-neuraminidase revealed | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dirr, L, El-Deeb, I, Guillon, P, Carroux, C, Chavas, L, von Itzstein, M. | | Deposit date: | 2015-01-08 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The catalytic mechanism of human parainfluenza virus type 3 haemagglutinin-neuraminidase revealed.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5M50
 
 | | Mechanism of microtubule minus-end recognition and protection by CAMSAP proteins | | Descriptor: | Calmodulin-regulated spectrin-associated protein 3, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Akhmanova, A, Moores, C.A, Baldus, M, Steinmetz, M.O, Topf, M, Roberts, A.J, Grant, B.J, Scarabelli, G, Joseph, A.-P, van Hooff, J.J.E, Houben, K, Hua, S, Luo, Y, Stangier, M.M, Jiang, K, Atherton, J. | | Deposit date: | 2016-10-20 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | A structural model for microtubule minus-end recognition and protection by CAMSAP proteins.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5IK0
 
 | | Tobacco 5-epi-aristolochene synthase with FPP | | Descriptor: | 5-epi-aristolochene synthase, FARNESYL DIPHOSPHATE, MAGNESIUM ION | | Authors: | Koo, H.J, Xu, Y, Louie, G.V, Bowman, M, Noel, J.P. | | Deposit date: | 2016-03-03 | | Release date: | 2016-10-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biosynthetic potential of sesquiterpene synthases: product profiles of Egyptian Henbane premnaspirodiene synthase and related mutants.
J.Antibiot., 69, 2016
|
|
5M4Q
 
 | | Crystal Structure of Wild-Type Human Prolidase with Mn ions and Pro ligand | | Descriptor: | GLYCEROL, HYDROXIDE ION, MANGANESE (II) ION, ... | | Authors: | Wilk, P, Weiss, M.S, Mueller, U, Dobbek, H. | | Deposit date: | 2016-10-18 | | Release date: | 2017-07-12 | | Last modified: | 2020-04-22 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Substrate specificity and reaction mechanism of human prolidase.
FEBS J., 284, 2017
|
|
7ALM
 
 | |
4XUW
 
 | | Structure of the hazelnut allergen, Cor a 8 | | Descriptor: | Non-specific lipid-transfer protein, PHOSPHATE ION | | Authors: | Offermann, L.R, Perdue, M.L, Bublin, M, Pfeifer, S, Dubiela, P, Hoffmann-Sommergruber, K, Chruszcz, M. | | Deposit date: | 2015-01-26 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural and Functional Characterization of the Hazelnut Allergen Cor a 8.
J.Agric.Food Chem., 63, 2015
|
|
