6O8N
 
 | |
4HUZ
 
 | | 2,6-Dichloro-p-hydroquinone 1,2-Dioxygenase | | Descriptor: | 2,6-dichloro-p-hydroquinone 1,2-dioxygenase, FE (III) ION, SULFATE ION | | Authors: | Hayes, R.P, Nissen, M.S, Green, A.R, Lewis, K.M, Xun, L, Kang, C. | | Deposit date: | 2012-11-05 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural characterization of 2,6-dichloro-p-hydroquinone 1,2-dioxygenase (PcpA) from Sphingobium chlorophenolicum, a new type of aromatic ring-cleavage enzyme.
Mol.Microbiol., 88, 2013
|
|
5Y48
 
 | | Crystal structure of the complex of Ribosome inactivating protein from Momordica balsamina with Pyrimidine-2,4-dione at 1.70 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Ribosome inactivating protein, URACIL | | Authors: | Singh, P.K, Pandey, S, Iqbal, N, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2017-08-01 | | Release date: | 2017-09-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Binding and structural studies of the complexes of type 1 ribosome inactivating protein from Momordica balsamina with uracil and uridine.
Proteins, 87, 2019
|
|
7YUY
 
 | | Structure of a mutated membrane-bound glycosyltransferase | | Descriptor: | (11R,14S)-17-amino-14-hydroxy-8,14-dioxo-9,13,15-trioxa-14lambda~5~-phosphaheptadecan-11-yl decanoate, 1,3-beta-glucan synthase component FKS1, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hu, X.L, Yang, P, Zhang, M, Liu, X.T, Yu, H.J. | | Deposit date: | 2022-08-18 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural and mechanistic insights into fungal beta-1,3-glucan synthase FKS1.
Nature, 616, 2023
|
|
5IHE
 
 | | D-family DNA polymerase - DP1 subunit (3'-5' proof-reading exonuclease) | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, ACETATE ION, ... | | Authors: | Sauguet, L, Raia, P, De Larue, M. | | Deposit date: | 2016-02-29 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Shared active site architecture between archaeal PolD and multi-subunit RNA polymerases revealed by X-ray crystallography.
Nat Commun, 7, 2016
|
|
5IJQ
 
 | | Crystal structure of autotaxin (ENPP2) re-refined | | Descriptor: | 7alpha-hydroxycholesterol, CALCIUM ION, Ectonucleotide pyrophosphatase/phosphodiesterase family member 2, ... | | Authors: | Hausmann, J, Joosten, R.P, Perrakis, A. | | Deposit date: | 2016-03-02 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural snapshots of the catalytic cycle of the phosphodiesterase Autotaxin.
J.Struct.Biol., 195, 2016
|
|
7AF2
 
 | | Salmonella typhimurium neuraminidase mutant (D62G) | | Descriptor: | GLYCEROL, PHOSPHATE ION, Sialidase | | Authors: | Salinger, M.T, Kuhn, P, Laver, W.G, Pape, T, Schneider, T.R, Sheldrick, G.M, Vimr, E.R, Garman, E.F. | | Deposit date: | 2020-09-19 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.792 Å) | | Cite: | Salmonella typhimurium neuraminidase mutant (D62G)
To Be Published
|
|
5IK6
 
 | | Tobacco 5-epi-aristolochene synthase with germacrene A and PPi | | Descriptor: | 5-epi-aristolochene synthase, DIPHOSPHATE, GERMACRENE A, ... | | Authors: | Koo, H.J, Xu, Y, Louie, G.V, Bowman, M, Noel, J.P. | | Deposit date: | 2016-03-03 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biosynthetic potential of sesquiterpene synthases: product profiles of Egyptian Henbane premnaspirodiene synthase and related mutants.
J.Antibiot., 69, 2016
|
|
4X2Y
 
 | | Crystal structure of a chimeric Murine Norovirus NS6 protease (inactive C139A mutant) in which the P4-P4 prime residues of the cleavage junction in the extended C-terminus have been replaced by the corresponding residues from the NS2-3 junction. | | Descriptor: | NS6 Protease,NS6 Protease | | Authors: | Leen, E.N, Pfeil, M.-P, Fernandes, H, Curry, S. | | Deposit date: | 2014-11-27 | | Release date: | 2015-02-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.417 Å) | | Cite: | Structure determination of Murine Norovirus NS6 proteases with C-terminal extensions designed to probe protease-substrate interactions.
Peerj, 3, 2015
|
|
5IM5
 
 | | Crystal structure of designed two-component self-assembling icosahedral cage I53-40 | | Descriptor: | Designed Keto-hydroxyglutarate-aldolase/keto-deoxy-phosphogluconate aldolase, Designed Riboflavin synthase | | Authors: | Liu, Y.A, Cascio, D, Sawaya, M.R, Bale, J.B, Collazo, M.J, Thomas, C, Sheffler, W, King, N.P, Baker, D, Yeates, T.O. | | Deposit date: | 2016-03-05 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.699 Å) | | Cite: | Accurate design of megadalton-scale two-component icosahedral protein complexes.
Science, 353, 2016
|
|
5YFW
 
 | | Crystal structure of ribose-1,5-bisphosphate isomerase mutant D204N from Pyrococcus horikoshii OT3 in complex with ribose-1,5-bisphosphate and AMP | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1,5-di-O-phosphono-alpha-D-ribofuranose, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Gogoi, P, Kanaujia, S.P. | | Deposit date: | 2017-09-22 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A presumed homologue of the regulatory subunits of eIF2B functions as ribose-1,5-bisphosphate isomerase in Pyrococcus horikoshii OT3.
Sci Rep, 8, 2018
|
|
6OSJ
 
 | |
4QKK
 
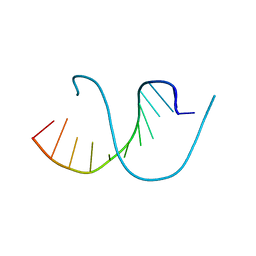 | | Crystal structure of an oligonucleotide containing 5-formylcytosine | | Descriptor: | DNA (5'-D(*CP*TP*AP*(5FC)P*GP*(5FC)P*GP*(5FC)P*GP*TP*AP*G)-3') | | Authors: | Raiber, E.-A, Murat, P, Chirgadze, D.Y, Luisi, B.F, Balasubramanian, S. | | Deposit date: | 2014-06-06 | | Release date: | 2014-12-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | 5-Formylcytosine alters the structure of the DNA double helix.
Nat.Struct.Mol.Biol., 22, 2015
|
|
6OKF
 
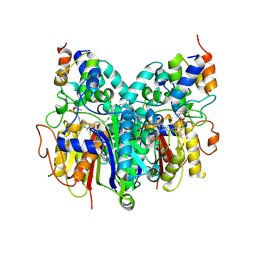 | | Crosslinked Crystal Structure of Type II Fatty Acid Synthase Ketosynthase, FabB, and C16-crypto Acyl Carrier Protein, AcpP | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 1, Acyl carrier protein, N-[2-(dodecanoylamino)ethyl]-N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alaninamide, ... | | Authors: | Mindrebo, J.T, Kim, W.E, Bartholow, T.G, Chen, A, Davis, T.D, La Clair, J, Burkart, M.D, Noel, J.P. | | Deposit date: | 2019-04-12 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Gating mechanism of elongating beta-ketoacyl-ACP synthases.
Nat Commun, 11, 2020
|
|
5I82
 
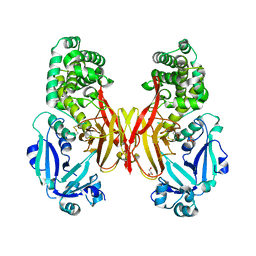 | | First Crystal Structure of E.coli Based Recombinant Diphtheria Toxin Mutant CRM197 | | Descriptor: | Diphtheria toxin, GLYCEROL, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Mishra, R.P.N, Goel, A, Dubrovska, I, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-02-18 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and immunological characterization of E. coli derived recombinant CRM 197 protein used as carrier in conjugate vaccines.
Biosci.Rep., 38, 2018
|
|
6V8U
 
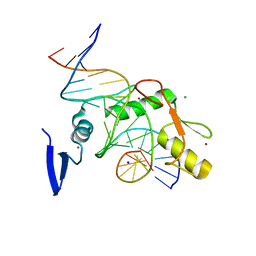 | | Kaiso (ZBTB33) zinc finger DNA binding domain in complex with a modified Kaiso binding sequence (KBS) | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*GP*TP*TP*AP*TP*TP*GP*GP*CP*AP*CP*GP*AP*AP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*TP*TP*CP*GP*TP*GP*CP*CP*AP*AP*TP*AP*AP*CP*G)-3'), ... | | Authors: | Nikolova, E.N, Stanfield, R.L, Dyson, H.J, Wright, P.E. | | Deposit date: | 2019-12-12 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | A Conformational Switch in the Zinc Finger Protein Kaiso Mediates Differential Readout of Specific and Methylated DNA Sequences.
Biochemistry, 59, 2020
|
|
5I9G
 
 | | Crystal structure of designed pentatricopeptide repeat protein dPPR-U8C2 in complex with its target RNA U8C2 | | Descriptor: | RNA (5'-R(*GP*GP*G*GP*UP*UP*UP*UP*CP*CP*UP*UP*UP*UP*CP*CP*CP*C)-3'), pentatricopeptide repeat protein dPPR-U8C2 | | Authors: | Shen, C, Zhang, D, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-02-20 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.288 Å) | | Cite: | Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins.
Nat Commun, 7, 2016
|
|
8U5F
 
 | | Crystal Structure of Trypsinized Clostridium perfringens Enterotoxin | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Heat-labile enterotoxin B chain, ... | | Authors: | Kapoor, S, Ogbu, C.P, Vecchio, A.J. | | Deposit date: | 2023-09-12 | | Release date: | 2023-09-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural Basis of Clostridium perfringens Enterotoxin Activation and Oligomerization by Trypsin.
Toxins, 15, 2023
|
|
5LJZ
 
 | | Structure of hantavirus envelope glycoprotein Gc in postfusion conformation | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Envelopment polyprotein, ... | | Authors: | Guardado-Calvo, P, Rey, F.A. | | Deposit date: | 2016-07-20 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mechanistic Insight into Bunyavirus-Induced Membrane Fusion from Structure-Function Analyses of the Hantavirus Envelope Glycoprotein Gc.
Plos Pathog., 12, 2016
|
|
5YG6
 
 | | Crystal structure of ribose-1,5-bisphosphate isomerase mutant C135S from Pyrococcus horikoshii OT3 in complex with ribose-1,5-bisphosphate and GMP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,5-di-O-phosphono-alpha-D-ribofuranose, GUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Gogoi, P, Kanaujia, S.P. | | Deposit date: | 2017-09-22 | | Release date: | 2018-02-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A presumed homologue of the regulatory subunits of eIF2B functions as ribose-1,5-bisphosphate isomerase in Pyrococcus horikoshii OT3.
Sci Rep, 8, 2018
|
|
6V8Z
 
 | |
4WYJ
 
 | | Adenovirus 3 head domain mutant V239D | | Descriptor: | Fiber protein, SULFATE ION | | Authors: | Lieber, A, Zubieta, C, Fender, P. | | Deposit date: | 2014-11-17 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Preclinical safety and efficacy studies with an affinity-enhanced epithelial junction opener and PEGylated liposomal doxorubicin.
Mol Ther Methods Clin Dev, 2, 2015
|
|
6VAB
 
 | | Mouse retromer sub-structure: VPS35/VPS35 flat dimer | | Descriptor: | Vacuolar protein sorting-associated protein 29, Vacuolar protein sorting-associated protein 35 | | Authors: | Kendall, A.K, Jackson, L.P. | | Deposit date: | 2019-12-17 | | Release date: | 2020-02-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Mammalian Retromer Is an Adaptable Scaffold for Cargo Sorting from Endosomes.
Structure, 28, 2020
|
|
5IBW
 
 | | Complex of MlcC bound to the tandem IQ motif of MyoC | | Descriptor: | Calcium-binding EF-hand domain-containing protein, Myosin IC heavy chain, SODIUM ION | | Authors: | Langelaan, D.N, Smith, S.P. | | Deposit date: | 2016-02-22 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Single-lobe Myosin Light Chain C in Complex with the Light Chain-binding Domains of Myosin-1C Provides Insights into Divergent IQ Motif Recognition.
J.Biol.Chem., 291, 2016
|
|
5LNV
 
 | | Crystal structure of Arabidopsis thaliana Pdx1-I320 complex from multiple crystals | | Descriptor: | (4~{S})-4-azanyl-5-oxidanyl-pent-1-en-3-one, PHOSPHATE ION, Pyridoxal 5'-phosphate synthase subunit PDX1.3, ... | | Authors: | Rodrigues, M.J, Windeisen, V, Zhang, Y, Guedez, G, Weber, S, Strohmeier, M, Hanes, J.W, Royant, A, Evans, G, Sinning, I, Ealick, S.E, Begley, T.P, Tews, I. | | Deposit date: | 2016-08-06 | | Release date: | 2017-01-18 | | Last modified: | 2018-09-19 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Lysine relay mechanism coordinates intermediate transfer in vitamin B6 biosynthesis.
Nat. Chem. Biol., 13, 2017
|
|
