7TYN
 
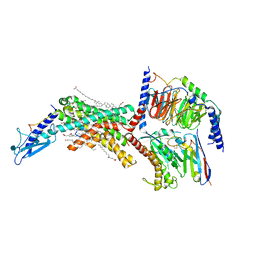 | | Calcitonin Receptor in complex with Gs and salmon calcitonin peptide | | Descriptor: | (2S)-2-{[(1R)-1-hydroxyhexadecyl]oxy}-3-{[(1R)-1-hydroxyoctadecyl]oxy}propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Cao, J, Belousoff, M.J, Johnson, R.M, Wootten, D.L, Sexton, P.M. | | Deposit date: | 2022-02-13 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | A structural basis for amylin receptor phenotype.
Science, 375, 2022
|
|
7LQD
 
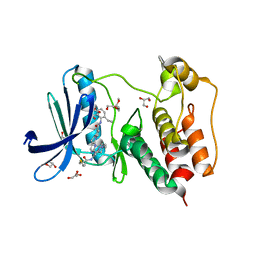 | | Structure of Human MPS1 (TTK) covalently bound to RMS-07 inhibitor | | Descriptor: | 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, Dual specificity protein kinase TTK or monopolar spindle 1 kinase, GLYCEROL, ... | | Authors: | Santiago, A.S, dos Reis, C.V, Serafim, R.A.M, Massirer, K.B, Arruda, P, Edwards, A.M, Counago, R.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-02-13 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Development of the First Covalent Monopolar Spindle Kinase 1 (MPS1/TTK) Inhibitor.
J.Med.Chem., 65, 2022
|
|
6OMY
 
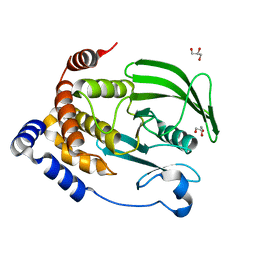 | | Protein Tyrosine Phosphatase 1B (1-301), P180A mutant, apo state | | Descriptor: | GLYCEROL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Cui, D.S, Lipchock, J.M, Loria, J.P. | | Deposit date: | 2019-04-19 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Uncovering the Molecular Interactions in the Catalytic Loop That Modulate the Conformational Dynamics in Protein Tyrosine Phosphatase 1B.
J.Am.Chem.Soc., 141, 2019
|
|
5G0H
 
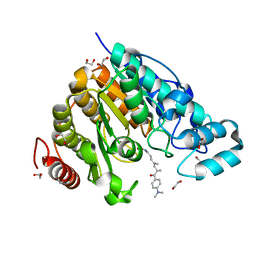 | | Crystal structure of Danio rerio HDAC6 CD2 in complex with (S)- trichostatin A | | Descriptor: | 1,2-ETHANEDIOL, HDAC6, POTASSIUM ION, ... | | Authors: | Miyake, Y, Keusch, J.J, Wang, L, Saito, M, Hess, D, Wang, X, Melancon, B.J, Helquist, P, Gut, H, Matthias, P. | | Deposit date: | 2016-03-18 | | Release date: | 2016-07-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Insights Into Hdac6 Tubulin Deacetylation and its Selective Inhibition
Nat.Chem.Biol., 12, 2016
|
|
7A93
 
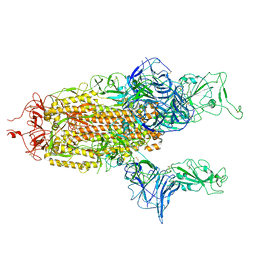 | |
7ZWG
 
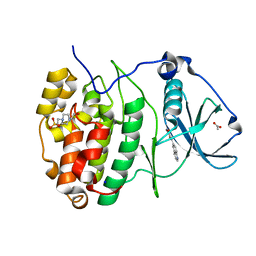 | | The Crystal structure of RO4493940 bound to CK2alpha | | Descriptor: | (5~{Z})-5-(quinolin-6-ylmethylidene)-1,3-thiazolidine-2,4-dione, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Brear, P, Hyvonen, M. | | Deposit date: | 2022-05-19 | | Release date: | 2023-05-31 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Chemical proteomics reveals the target landscape of 1,000 kinase inhibitors.
Nat.Chem.Biol., 20, 2024
|
|
7ZWE
 
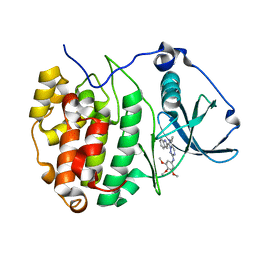 | | The Crystal structure of GW8695 bound to CK2alpha | | Descriptor: | 7-(1~{H}-indol-2-yl)-5-methyl-~{N}-(3,4,5-trimethoxyphenyl)imidazo[5,1-f][1,2,4]triazin-2-amine, Casein kinase II subunit alpha | | Authors: | Brear, P, Hyvonen, M. | | Deposit date: | 2022-05-19 | | Release date: | 2023-05-31 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Chemical proteomics reveals the target landscape of 1,000 kinase inhibitors.
Nat.Chem.Biol., 20, 2024
|
|
8DI4
 
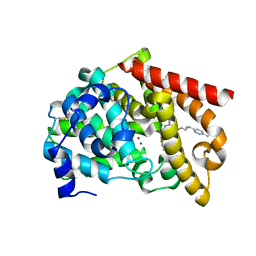 | | Discovery of MK-8189, a highly potent and selective PDE10A inhibitor for the treatment of schizophrenia | | Descriptor: | 2-methyl-6-{[(1S,2S)-2-(5-methylpyridin-2-yl)cyclopropyl]methoxy}-N-[(5-methyl-1,3,4-thiadiazol-2-yl)methyl]pyrimidin-4-amine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Hayes, R.P, Yan, Y. | | Deposit date: | 2022-06-28 | | Release date: | 2023-02-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.016 Å) | | Cite: | Discovery of MK-8189, a Highly Potent and Selective PDE10A Inhibitor for the Treatment of Schizophrenia.
J.Med.Chem., 66, 2023
|
|
7JJD
 
 | |
6UHQ
 
 | | Crystal Structure of C148 mGFP-cDNA-3 | | Descriptor: | C148 mGFP-cDNA-3, UNKNOWN LIGAND | | Authors: | Winegar, P.W, Hayes, O.G, McMillan, J.R, Figg, C.A, Focia, P.J, Mirkin, C.A. | | Deposit date: | 2019-09-27 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | DNA-Directed Protein Packing within Single Crystals.
Chem, 6, 2020
|
|
7QB2
 
 | | Pim1 in complex with (E)-4-((6-amino-1-methyl-2-oxoindolin-3-ylidene)methyl)benzoic acid and Pimtide | | Descriptor: | 4-[(E)-(6-azanyl-1-methyl-2-oxidanylidene-indol-3-ylidene)methyl]benzoic acid, GLYCEROL, Pimtide, ... | | Authors: | Hochban, P.M.M, Heine, A, Diederich, W.E. | | Deposit date: | 2021-11-18 | | Release date: | 2022-11-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Pose, duplicate, then elaborate: Steps towards increased affinity for inhibitors targeting the specificity surface of the Pim-1 kinase.
Eur.J.Med.Chem., 245, 2023
|
|
6KM8
 
 | | Crystal Structure of Momordica charantia 7S globulin | | Descriptor: | 7S globulin, ACETATE ION, COPPER (II) ION | | Authors: | Kesari, P, Pratap, S, Dhankhar, P, Dalal, V, Kumar, P. | | Deposit date: | 2019-07-31 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.099 Å) | | Cite: | Structural characterization and in-silico analysis of Momordica charantia 7S globulin for stability and ACE inhibition.
Sci Rep, 10, 2020
|
|
8PJE
 
 | | Human Leukocyte Antigen class II allotype DR1 presenting influenza A virus haemagglutinin (HA)306-318 PKYVKQNTLKLAT | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | MacLachlan, B.J, Wall, A, Greenshields-Watson, A.L, Hesketh, S.J, Cole, D.K, Rizkallah, P.J, Godkin, A.J. | | Deposit date: | 2023-06-23 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A targeted single mutation in influenza A virus universal epitope transforms immunogenicity and protective immunity via CD4 + T cell activation.
Cell Rep, 43, 2024
|
|
8PJF
 
 | | Human Leukocyte Antigen class II allotype DR1 presenting P11T->R modified influenza A virus haemagglutinin (HA)306-318 PKYVKQNTLKLAR | | Descriptor: | 1,2-ETHANEDIOL, HLA class II histocompatibility antigen, DR alpha chain, ... | | Authors: | MacLachlan, B.J, Wall, A, Greenshields-Watson, A.L, Cole, D.K, Rizkallah, P.J, Godkin, A.J. | | Deposit date: | 2023-06-23 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | A targeted single mutation in influenza A virus universal epitope transforms immunogenicity and protective immunity via CD4 + T cell activation.
Cell Rep, 43, 2024
|
|
7AAJ
 
 | | Human porphobilinogen deaminase in complex with cofactor | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, GLYCEROL, Porphobilinogen deaminase | | Authors: | Kallio, J.P, Bustad, H.J, Martinez, A. | | Deposit date: | 2020-09-04 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of porphobilinogen deaminase mutants reveals that arginine-173 is crucial for polypyrrole elongation mechanism.
Iscience, 24, 2021
|
|
7TOG
 
 | | Crystal structure of carbohydrate esterase PbeAcXE, apoenzyme | | Descriptor: | SGNH hydrolase | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Jurak, E, Master, E. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-13 | | Last modified: | 2022-11-02 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Elucidating Sequence and Structural Determinants of Carbohydrate Esterases for Complete Deacetylation of Substituted Xylans.
Molecules, 27, 2022
|
|
7TOI
 
 | | Crystal structure of carbohydrate esterase PbeAcXE, in complex with acetate | | Descriptor: | ACETATE ION, SGNH hydrolase | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Jurak, E, Master, E. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-13 | | Last modified: | 2022-11-02 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Elucidating Sequence and Structural Determinants of Carbohydrate Esterases for Complete Deacetylation of Substituted Xylans.
Molecules, 27, 2022
|
|
7QFM
 
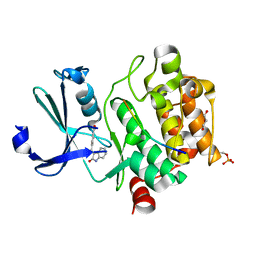 | | Pim1 in complex with (E)-4-((2-oxoindolin-3-ylidene)methyl)benzoic acid and Pimtide | | Descriptor: | 4-[(~{E})-(2-oxidanylidene-1~{H}-indol-3-ylidene)methyl]benzoic acid, GLYCEROL, Pimtide, ... | | Authors: | Hochban, P.M.M, Heine, A, Diederich, W.E. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pose, duplicate, then elaborate: Steps towards increased affinity for inhibitors targeting the specificity surface of the Pim-1 kinase.
Eur.J.Med.Chem., 245, 2023
|
|
7TOH
 
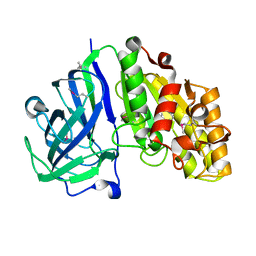 | | Crystal structure of carbohydrate esterase PbeAcXE, in complex with MeGlcpA-Xylp | | Descriptor: | 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-beta-D-xylopyranose, SGNH hydrolase | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Jurak, E, Master, E. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-13 | | Last modified: | 2022-11-02 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Elucidating Sequence and Structural Determinants of Carbohydrate Esterases for Complete Deacetylation of Substituted Xylans.
Molecules, 27, 2022
|
|
5FPT
 
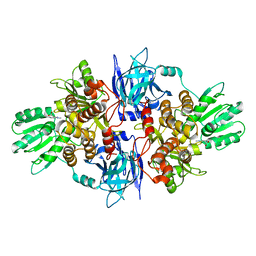 | | Structure of hepatitis C virus (HCV) full-length NS3 complex with small-molecule ligand 2-(1-methyl-1H-indol-3-yl)acetic acid (AT3437) in an alternate binding site. | | Descriptor: | (1-methyl-1H-indol-3-yl)acetic acid, HEPATITIS C VIRUS FULL-LENGTH NS3 COMPLEX | | Authors: | Jhoti, H, Ludlow, R.F, Saini, H.K, Tickle, I.J, Verdonk, M, Pathuri, P, Williams, P.A. | | Deposit date: | 2015-12-02 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8PJG
 
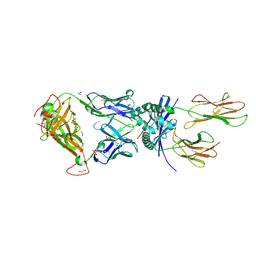 | | F11 TCR in complex with Human Leukocyte Antigen class II allotype DR1 presenting P11T->R modified influenza A virus haemagglutinin (HA)306-318 PKYVKQNTLKLAR | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, HLA class II histocompatibility antigen, ... | | Authors: | MacLachlan, B.J, Wall, A, Greenshields-Watson, A.L, Cole, D.K, Rizkallah, P.J, Godkin, A.J. | | Deposit date: | 2023-06-23 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | A targeted single mutation in influenza A virus universal epitope transforms immunogenicity and protective immunity via CD4 + T cell activation.
Cell Rep, 43, 2024
|
|
1MQ3
 
 | | Human DNA Polymerase Beta Complexed With Gapped DNA Containing an 8-oxo-7,8-dihydro-Guanine Template Paired with dCTP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*CP*CP*GP*AP*CP*(8OG)P*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(DOC))-3', ... | | Authors: | Krahn, J.M, Beard, W.A, Miller, H, Grollman, A.P, Wilson, S.H. | | Deposit date: | 2002-09-13 | | Release date: | 2003-01-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of DNA Polymerase beta with the Mutagenic DNA Lesion 8-oxodeoxyguanine Reveals
Structural Insights into its Coding Potential
Structure, 11, 2003
|
|
8F7Z
 
 | | VRC34.01_mm28 bound to fusion peptide | | Descriptor: | HIV-1 Env Fusion Peptide, VRC34_m228 Light Chain, VRC34_mm28 Heavy Chain | | Authors: | Olia, A.S, Kwong, P.D. | | Deposit date: | 2022-11-21 | | Release date: | 2023-11-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Antibody-directed evolution reveals a mechanism for enhanced neutralization at the HIV-1 fusion peptide site.
Nat Commun, 14, 2023
|
|
4XMK
 
 | |
7AAK
 
 | | Human porphobilinogen deaminase R173W mutant crystallized in the ES2 intermediate state | | Descriptor: | 3-[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-3-(3-hydroxy-3-oxopropyl)-5-methyl-1~{H}-pyrrol-2-yl]methyl]-3-(3-hydroxy-3-oxopropyl)-1~{H}-pyrrol-2-yl]methyl]-3-(3-hydroxy-3-oxopropyl)-1~{H}-pyrrol-2-yl]methyl]-1~{H}-pyrrol-3-yl]propanoic acid, GLYCEROL, Porphobilinogen deaminase | | Authors: | Kallio, J.P, Bustad, H.J, Martinez, A. | | Deposit date: | 2020-09-04 | | Release date: | 2021-02-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterization of porphobilinogen deaminase mutants reveals that arginine-173 is crucial for polypyrrole elongation mechanism.
Iscience, 24, 2021
|
|
