5HU0
 
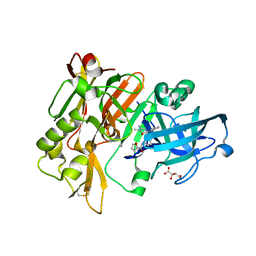 | | BACE1 in complex with 4-(3-(furan-2-carboxamido)phenyl)-1-methyl-5-oxo-4-phenylimidazolidin-2-iminium | | Descriptor: | Beta-secretase 1, L(+)-TARTARIC ACID, N-{3-[(2E,4R)-2-imino-1-methyl-5-oxo-4-phenylimidazolidin-4-yl]phenyl}furan-2-carboxamide | | Authors: | Orth, P. | | Deposit date: | 2016-01-27 | | Release date: | 2016-11-09 | | Last modified: | 2017-01-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of the 3-Imino-1,2,4-thiadiazinane 1,1-Dioxide Derivative Verubecestat (MK-8931)-A beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 Inhibitor for the Treatment of Alzheimer's Disease.
J. Med. Chem., 59, 2016
|
|
6TAE
 
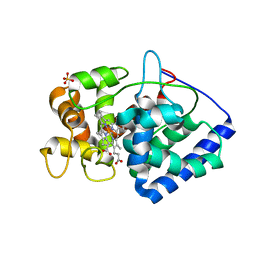 | | Neutron structure of ferric ascorbate peroxidase | | Descriptor: | Ascorbate peroxidase, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Kwon, H, Basran, J, Devos, J.M, Schrader, T.E, Ostermann, A, Blakeley, M.P, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2019-10-29 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (1.9 Å), X-RAY DIFFRACTION | | Cite: | Visualizing the protons in a metalloenzyme electron proton transfer pathway.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6TE9
 
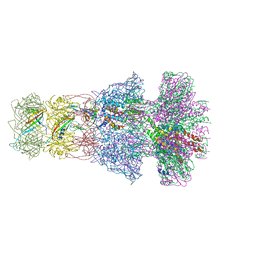 | | Neck of native GTA particle computed with C6 symmetry | | Descriptor: | Adaptor protein Rcc01688, Phage major tail protein, TP901-1 family, ... | | Authors: | Bardy, P, Fuzik, T, Hrebik, D, Pantucek, R, Beatty, J.T, Plevka, P. | | Deposit date: | 2019-11-11 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structure and mechanism of DNA delivery of a gene transfer agent.
Nat Commun, 11, 2020
|
|
6TFZ
 
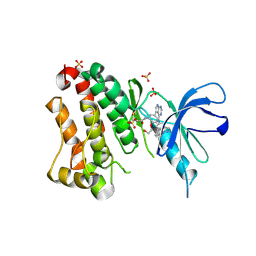 | | Crystal Structure of EGFR T790M/V948R in Complex with Covalent Pyrrolopyrimidine 19 | | Descriptor: | 1,2-ETHANEDIOL, Epidermal growth factor receptor, SULFATE ION, ... | | Authors: | Niggenaber, J, Mueller, M.P, Rauh, D. | | Deposit date: | 2019-11-14 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Targeting Her2-insYVMA with Covalent Inhibitors-A Focused Compound Screening and Structure-Based Design Approach.
J.Med.Chem., 63, 2020
|
|
4V3O
 
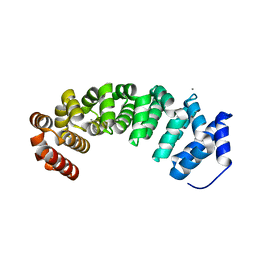 | | Designed armadillo repeat protein with 5 internal repeats, 2nd generation C-cap and 3rd generation N-cap. | | Descriptor: | ACETATE ION, CALCIUM ION, YIII_M5_AII | | Authors: | Reichen, C, Madhurantakam, C, Pluckthun, A, Mittl, P. | | Deposit date: | 2014-10-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Designed Armadillo-Repeat Proteins Show Propagation of Inter-Repeat Interface Effects
Acta Crystallogr.,Sect.D, 72, 2016
|
|
1GG9
 
 | | CRYSTAL STRUCTURE OF CATALASE HPII FROM ESCHERICHIA COLI, HIS128ASN VARIANT. | | Descriptor: | CATALASE HPII, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Melik-Adamyan, W.R, Bravo, J, Carpena, X, Switala, J, Mate, M.J, Fita, I, Loewen, P.C. | | Deposit date: | 2000-08-11 | | Release date: | 2000-08-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Substrate flow in catalases deduced from the crystal structures of active site variants of HPII from Escherichia coli.
Proteins, 44, 2001
|
|
6TE8
 
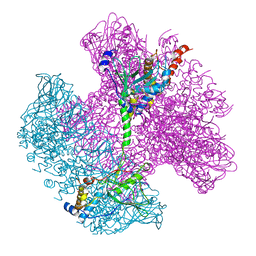 | | Neck of native GTA particle computed with C12 symmetry | | Descriptor: | Adaptor protein Rcc01688, Phage portal protein, HK97 family | | Authors: | Bardy, P, Fuzik, T, Hrebik, D, Pantucek, R, Beatty, J.T, Plevka, P. | | Deposit date: | 2019-11-11 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structure and mechanism of DNA delivery of a gene transfer agent.
Nat Commun, 11, 2020
|
|
4V5W
 
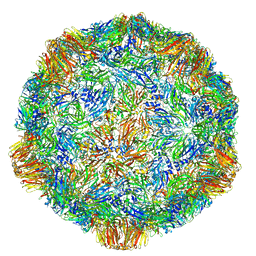 | | Grapevine Fanleaf virus | | Descriptor: | COAT PROTEIN | | Authors: | Schellenberger, P, Demangeat, G, Ritzenthaler, C, Lorber, B, Sauter, C. | | Deposit date: | 2011-05-10 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Exploiting Protein Engineering and Crystal Polymorphism for Successful X-Ray Structure Determination
Cryst.Growth Des., 11, 2011
|
|
6TOA
 
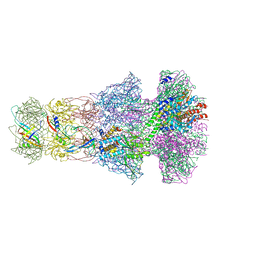 | | Neck of empty GTA particle computed with C6 symmetry | | Descriptor: | Adaptor protein Rcc01688, Portal protein Rcc01684, Stopper protein Rcc01689, ... | | Authors: | Bardy, P, Fuzik, T, Hrebik, D, Pantucek, R, Beatty, J.T, Plevka, P. | | Deposit date: | 2019-12-11 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structure and mechanism of DNA delivery of a gene transfer agent.
Nat Commun, 11, 2020
|
|
5HUG
 
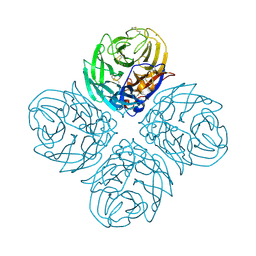 | | The crystal structure of neuraminidase from A/American green-winged teal/Washington/195750/2014 influenza virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Yang, H, Carney, P.J, Guo, Z, Chang, J.C, Stevens, J. | | Deposit date: | 2016-01-27 | | Release date: | 2016-04-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular Characterizations of Surface Proteins Hemagglutinin and Neuraminidase from Recent H5Nx Avian Influenza Viruses.
J.Virol., 90, 2016
|
|
4ZPV
 
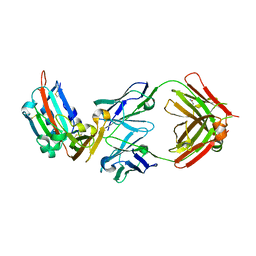 | | Structure of MERS-Coronavirus Spike Receptor-binding Domain (England1 Strain) in Complex with Vaccine-Elicited Murine Neutralizing Antibody D12 (Crystal Form 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D12 Fab Heavy chain, D12 Fab light chain, ... | | Authors: | Joyce, M.G, Mascola, J.R, Graham, B.S, Kwong, P.D. | | Deposit date: | 2015-05-08 | | Release date: | 2015-10-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Evaluation of candidate vaccine approaches for MERS-CoV.
Nat Commun, 6, 2015
|
|
4W90
 
 | | Crystal structure of Bacillus subtilis cyclic-di-AMP riboswitch ydaO | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, MAGNESIUM ION, U1 small nuclear ribonucleoprotein A, ... | | Authors: | Jones, C.P, Ferre-D'Amare, A.R. | | Deposit date: | 2014-08-26 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.118 Å) | | Cite: | Crystal structure of a c-di-AMP riboswitch reveals an internally pseudo-dimeric RNA.
Embo J., 33, 2014
|
|
5HYP
 
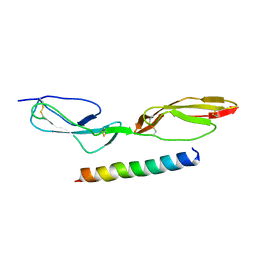 | |
4NR5
 
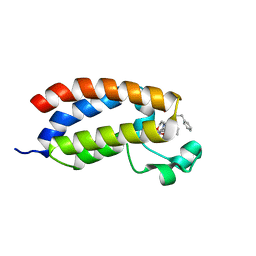 | | Crystal structure of the bromodomain of human CREBBP in complex with an isoxazolyl-benzimidazole ligand | | Descriptor: | 1,2-ETHANEDIOL, 5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[2-(morpholin-4-yl)ethyl]-2-(2-phenylethyl)-1H-benzimidazole, CREB-binding protein, ... | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Hay, D, Fedorov, O, Martin, S, Pike, A.W, von Delft, F, Brennan, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-11-26 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of the bromodomain of human CREBBP in complex with an isoxazolyl-benzimidazole ligand
To be Published
|
|
4UU1
 
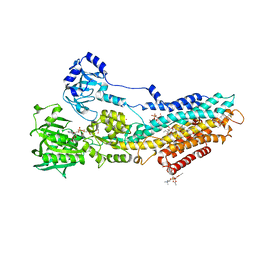 | | CRYSTAL STRUCTURE OF (SR) CALCIUM-ATPASE E2(TG) IN THE PRESENCE OF DOPC | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Drachmann, N.D, Olesen, C, Moeller, J.V, Guo, Z, Nissen, P, Bublitz, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Comparing Crystal Structures of Ca(2+) -ATPase in the Presence of Different Lipids.
FEBS J., 281, 2014
|
|
4W8Q
 
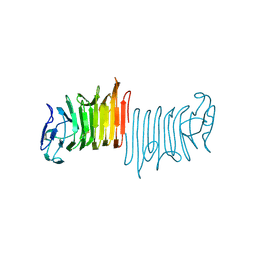 | |
4WD6
 
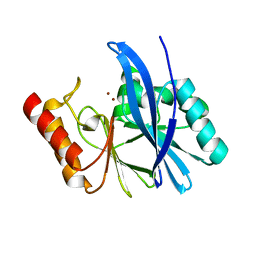 | |
5HTZ
 
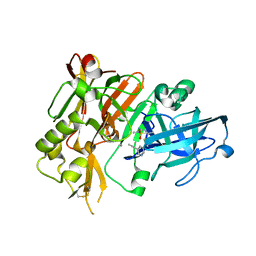 | | BACE1 in complex with (S)-5-(3-chloro-5-(5-(prop-1-yn-1-yl)pyridin-3-yl)thiophen-2-yl)-2,5-dimethyl-1,2,4-thiadiazinan-3-iminium 1,1-dioxide | | Descriptor: | (3E,5S)-5-{3-chloro-5-[5-(prop-1-yn-1-yl)pyridin-3-yl]thiophen-2-yl}-2,5-dimethyl-1,2,4-thiadiazinan-3-imine 1,1-dioxide, Beta-secretase 1 | | Authors: | Orth, P. | | Deposit date: | 2016-01-27 | | Release date: | 2016-11-09 | | Last modified: | 2017-01-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of the 3-Imino-1,2,4-thiadiazinane 1,1-Dioxide Derivative Verubecestat (MK-8931)-A beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 Inhibitor for the Treatment of Alzheimer's Disease.
J. Med. Chem., 59, 2016
|
|
3FQ5
 
 | |
2Q9L
 
 | | Crystal structure of iMazG from Vibrio DAT 722: Ctag-iMazG (P43212) | | Descriptor: | Hypothetical protein, MAGNESIUM ION | | Authors: | Robinson, A, Guilfoyle, A.P, Harrop, S.J, Boucher, Y, Stokes, H.W, Curmi, P.M.G, Mabbutt, B.C. | | Deposit date: | 2007-06-13 | | Release date: | 2007-10-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A putative house-cleaning enzyme encoded within an integron array: 1.8 A crystal structure defines a new MazG subtype.
Mol.Microbiol., 66, 2007
|
|
4UZV
 
 | | Structure of a triple mutant of ASV-TfTrHb | | Descriptor: | ACETATE ION, HEMOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Baiocco, P, Bonamore, A, Sciamanna, N, Ilari, A, Boechi, L, Boffi, A, Smulevich, G, Feis, A. | | Deposit date: | 2014-09-09 | | Release date: | 2014-09-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Role of Local Structure and Dynamics of Small Ligand Migration in Proteins: A Study of a Mutated Truncated Hemoprotein from Thermobifida Fusca by Time Resolved Mir Spectroscopy.
J.Phys.Chem.B, 118, 2014
|
|
1UM4
 
 | | Catalytic Antibody 21H3 with hapten | | Descriptor: | (1R)-1-PHENYLETHYL 4-(ACETYLAMINO)BENZYLPHOSPHONATE, Antibody 21H3 H chain, Antibody 21H3 L chain | | Authors: | Beuscher IV, A.E, Reuter, J, Olson, A.J, Romesberg, F.E, Schultz, P.G, Wirsching, P, Janda, K.D, Lerner, R.A, Stevens, R.C. | | Deposit date: | 2003-09-23 | | Release date: | 2004-10-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Studies of an Efficient Catalytic Antibody Operating by Ping-Pong and Induced Fit Mechanisms
To be Published
|
|
4NDZ
 
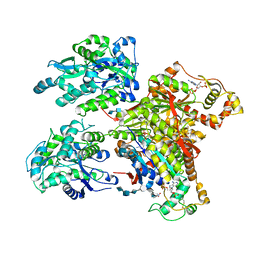 | | Structure of Maltose Binding Protein fusion to 2-O-Sulfotransferase with bound heptasaccharide and PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Maltose-binding periplasmic protein, Heparan sulfate 2-O-sulfotransferase 1 fusion, ... | | Authors: | Liu, C, Sheng, J, Krahn, J.M, Perera, L, Xu, Y, Hsieh, P, Liu, J, Pedersen, L.C. | | Deposit date: | 2013-10-28 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Deciphering the role of 2-O-sulfotransferase in regulating heparan sulfate biosynthesis
To be Published
|
|
5HO7
 
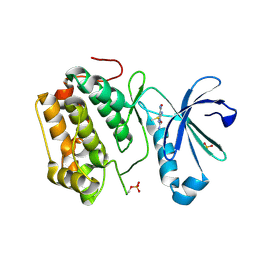 | | DISCOVERY OF NOVEL 7-AZAINDOLES AS PDK1 INHIBITORS | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, 5-amino-3-(methylsulfanyl)-1H-pyrazole-1,4-dicarboxamide, SULFATE ION | | Authors: | Wucherer-Plietker, M, Esdar, C, Knoechel, T, Hillertz, P, Heinrich, T, Buchstaller, H.P, Greiner, H, Dorsch, D, Calderini, M, Bruge, D, Mueller, T.J.J, Graedler, U. | | Deposit date: | 2016-01-19 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of novel 7-azaindoles as PDK1 inhibitors.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
3FRL
 
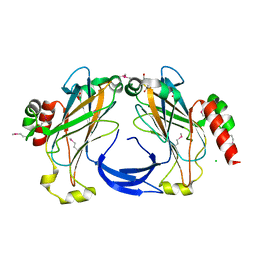 | | The 2.25 A crystal structure of LipL32, the major surface antigen of Leptospira interrogans serovar Copenhageni | | Descriptor: | 2,2',2''-NITRILOTRIETHANOL, CHLORIDE ION, LipL32, ... | | Authors: | Farah, C.S, Guzzo, C.R, Hauk, P, Ho, P.L. | | Deposit date: | 2009-01-08 | | Release date: | 2009-06-16 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure and calcium-binding activity of LipL32, the major surface antigen of pathogenic Leptospira sp.
J.Mol.Biol., 390, 2009
|
|
