6WCU
 
 | | Crystal structure of coiled coil region of human septin 5 | | Descriptor: | Septin-5 | | Authors: | Cabrejos, D.A.L, Cavini, I, Sala, F.A, Valadares, N.F, Pereira, H.M, Brandao-Neto, J, Nascimento, A.F.Z, Uson, I, Araujo, A.P.U, Garratt, R.C. | | Deposit date: | 2020-03-31 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Orientational Ambiguity in Septin Coiled Coils and its Structural Basis.
J.Mol.Biol., 433, 2021
|
|
3HSH
 
 | |
6WSM
 
 | | Crystal structure of coiled coil region of human septin 8 | | Descriptor: | SULFATE ION, Septin-8 | | Authors: | Cabrejos, D.A.L, Cavini, I, Sala, F.A, Valadares, N.F, Pereira, H.M, Brandao-Neto, J, Nascimento, A.F.Z, Uson, I, Araujo, A.P.U, Garratt, R.C. | | Deposit date: | 2020-05-01 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.451 Å) | | Cite: | Orientational Ambiguity in Septin Coiled Coils and its Structural Basis.
J.Mol.Biol., 433, 2021
|
|
1NGM
 
 | | Crystal structure of a yeast Brf1-TBP-DNA ternary complex | | Descriptor: | 5'-D(*AP*AP*AP*AP*AP*AP*CP*AP*TP*TP*TP*TP*TP*TP*TP*AP*TP*AP*G)-3', 5'-D(*CP*TP*AP*TP*AP*AP*AP*AP*AP*AP*AP*TP*GP*TP*TP*TP*TP*TP*T)-3', Transcription factor IIIB BRF1 subunit, ... | | Authors: | Juo, Z.S, Kassavetis, G.A, Wang, J, Geiduschek, E.P, Sigler, P.B. | | Deposit date: | 2002-12-17 | | Release date: | 2003-03-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of a transcription factor IIIB core interface ternary complex
Nature, 422, 2003
|
|
7A93
 
 | |
7RNQ
 
 | |
6UHN
 
 | | Crystal Structure of C148 mGFP-cDNA-1 | | Descriptor: | C148 mGFP-cDNA-1, UNKNOWN LIGAND | | Authors: | Winegar, P.W, Hayes, O.G, McMillan, J.R, Figg, C.A, Focia, P.J, Mirkin, C.A. | | Deposit date: | 2019-09-27 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | DNA-Directed Protein Packing within Single Crystals.
Chem, 6, 2020
|
|
8GOG
 
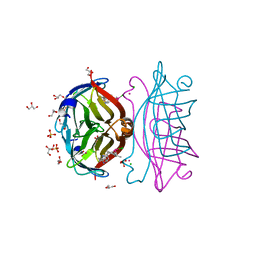 | | Structure of streptavidin mutant (S112Y-K121E) complexed with biotin-cyclopentadienyl-rhodium (III)(Cp*-Rh(III)) | | Descriptor: | CHLORIDE ION, GLYCEROL, RHODIUM(III) ION, ... | | Authors: | Sairaman, A, Mukherjee, P, Maiti, D, Bhaumik, P. | | Deposit date: | 2022-08-24 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enantiodivergent synthesis of isoindolones catalysed by a Rh(III)-based artificial metalloenzyme
Nat Synth, 2024
|
|
7AAJ
 
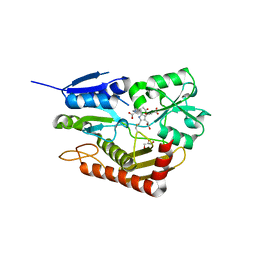 | | Human porphobilinogen deaminase in complex with cofactor | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, GLYCEROL, Porphobilinogen deaminase | | Authors: | Kallio, J.P, Bustad, H.J, Martinez, A. | | Deposit date: | 2020-09-04 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of porphobilinogen deaminase mutants reveals that arginine-173 is crucial for polypyrrole elongation mechanism.
Iscience, 24, 2021
|
|
6WTA
 
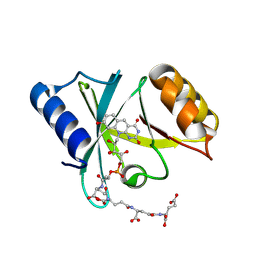 | |
6U3Q
 
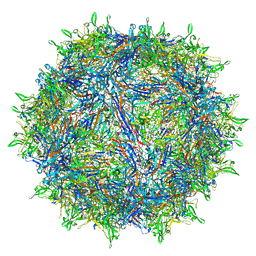 | | The atomic structure of a human adeno-associated virus capsid isolate (AAVhu69/AAVv66) | | Descriptor: | Capsid protein VP1 | | Authors: | Hsu, H.-L, Brown, A, Loveland, A, Tai, P, Korostelev, A, Gao, G. | | Deposit date: | 2019-08-22 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Structural characterization of a novel human adeno-associated virus capsid with neurotropic properties.
Nat Commun, 11, 2020
|
|
7AAK
 
 | | Human porphobilinogen deaminase R173W mutant crystallized in the ES2 intermediate state | | Descriptor: | 3-[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-3-(3-hydroxy-3-oxopropyl)-5-methyl-1~{H}-pyrrol-2-yl]methyl]-3-(3-hydroxy-3-oxopropyl)-1~{H}-pyrrol-2-yl]methyl]-3-(3-hydroxy-3-oxopropyl)-1~{H}-pyrrol-2-yl]methyl]-1~{H}-pyrrol-3-yl]propanoic acid, GLYCEROL, Porphobilinogen deaminase | | Authors: | Kallio, J.P, Bustad, H.J, Martinez, A. | | Deposit date: | 2020-09-04 | | Release date: | 2021-02-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterization of porphobilinogen deaminase mutants reveals that arginine-173 is crucial for polypyrrole elongation mechanism.
Iscience, 24, 2021
|
|
8BUV
 
 | | HIV-1 Integrase Catalytic Core Domain and C-Terminal Domain in Complex with Allosteric Integrase Inhibitor LEDGIN 3 | | Descriptor: | (6-chloro-2-oxo-4-phenyl-1,2-dihydroquinolin-3-yl)acetic acid, 1,2-ETHANEDIOL, Integrase, ... | | Authors: | Singer, M.R, Pye, V.E, Cook, N.J, Cherepanov, P. | | Deposit date: | 2022-11-30 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | HIV-1 Integrase Catalytic Core Domain and C-Terminal Domain in Complex with Allosteric Integrase Inhibitor LEDGIN 3
To Be Published
|
|
6OPH
 
 | | phosphorylated ERK2 with GDC-0994 | | Descriptor: | 1-[(1~{S})-1-(4-chloranyl-3-fluoranyl-phenyl)-2-oxidanyl-ethyl]-4-[2-[(2-methylpyrazol-3-yl)amino]pyrimidin-4-yl]pyridin-2-one, Mitogen-activated protein kinase 1 | | Authors: | Vigers, G.P, Smith, D. | | Deposit date: | 2019-04-25 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Activation loop dynamics are controlled by conformation-selective inhibitors of ERK2.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6ZPA
 
 | | Cyanophage S-2L HD phosphohydrolase (DatZ) bound to dA and one catalytic Zn2+ ion | | Descriptor: | (2R,3S,5R)-5-(6-amino-9H-purin-9-yl)-tetrahydro-2-(hydroxymethyl)furan-3-ol, DatZ, LITHIUM ION, ... | | Authors: | Czernecki, D, Legrand, P, Delarue, M. | | Deposit date: | 2020-07-08 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (0.86000258 Å) | | Cite: | How cyanophage S-2L rejects adenine and incorporates 2-aminoadenine to saturate hydrogen bonding in its DNA.
Nat Commun, 12, 2021
|
|
6ZPC
 
 | | Cyanophage S-2L HD phosphohydrolase (DatZ) bound to dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DatZ, LITHIUM ION, ... | | Authors: | Czernecki, D, Legrand, P, Delarue, M. | | Deposit date: | 2020-07-08 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.2683593 Å) | | Cite: | How cyanophage S-2L rejects adenine and incorporates 2-aminoadenine to saturate hydrogen bonding in its DNA.
Nat Commun, 12, 2021
|
|
6ZPB
 
 | | Cyanophage S-2L HD phosphohydrolase (DatZ) bound to dA and two catalytic Co2+ ions | | Descriptor: | (2R,3S,5R)-5-(6-amino-9H-purin-9-yl)-tetrahydro-2-(hydroxymethyl)furan-3-ol, COBALT (II) ION, DatZ | | Authors: | Czernecki, D, Legrand, P, Delarue, M. | | Deposit date: | 2020-07-08 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.72097385 Å) | | Cite: | How cyanophage S-2L rejects adenine and incorporates 2-aminoadenine to saturate hydrogen bonding in its DNA.
Nat Commun, 12, 2021
|
|
8SRX
 
 | | Crystal structure of BAK-BAX heterodimer with lysoPC | | Descriptor: | 1,2-ETHANEDIOL, Apoptosis regulator BAX, Bcl-2 homologous antagonist/killer, ... | | Authors: | Brouwer, J.M, Czabotar, P.E, Colman, P.M, Miller, M.S. | | Deposit date: | 2023-05-07 | | Release date: | 2023-12-27 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Sequence differences between BAX and BAK core domains manifest as differences in their interactions with lipids.
Febs J., 291, 2024
|
|
8BPP
 
 | |
8B8I
 
 | |
5DX1
 
 | |
6ZZ5
 
 | | Structure of soluble SmhB of the tripartite alpha-pore forming toxin, Smh, from Serratia marcescens. | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, SmhB | | Authors: | Churchill-Angus, A.M, Baker, P.J. | | Deposit date: | 2020-08-03 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Characterisation of a tripartite alpha-pore forming toxin from Serratia marcescens
Sci Rep, 11, 2021
|
|
5HYT
 
 | |
7A26
 
 | |
6OJX
 
 | |
