8AKU
 
 | | 250 A SynPspA rod after incubation with ATP | | Descriptor: | Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Non-canonical ATPase activity drives PspA membrane constriction
To Be Published
|
|
8AKY
 
 | | 290 A SynPspA rod after incubation with ATP | | Descriptor: | Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Non-canonical ATPase activity drives PspA membrane constriction
To Be Published
|
|
8AKZ
 
 | | 320 A SynPspA rod after incubation with ATP | | Descriptor: | Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Non-canonical ATPase activity drives PspA membrane constriction
To Be Published
|
|
6ZEK
 
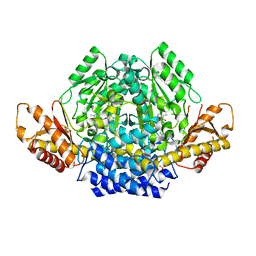 | | Crystal structure of mouse CSAD | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Cysteine sulfinic acid decarboxylase, ... | | Authors: | Mahootchi, E, Raasakka, A, Haavik, J, Kursula, P. | | Deposit date: | 2020-06-16 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and substrate specificity determinants of the taurine biosynthetic enzyme cysteine sulphinic acid decarboxylase.
J.Struct.Biol., 213, 2021
|
|
7RMK
 
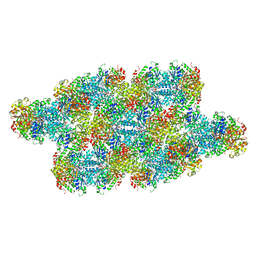 | | Yeast CTP Synthase (Ura7) Bundle bound to substrates at low pH | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CTP synthase, URIDINE 5'-TRIPHOSPHATE | | Authors: | Hansen, J.M, Lynch, E.M, Farrell, D.P, DiMaio, F, Quispe, J, Kollman, J.M. | | Deposit date: | 2021-07-27 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Cryo-EM structures of CTP synthase filaments reveal mechanism of pH-sensitive assembly during budding yeast starvation.
Elife, 10, 2021
|
|
8AL0
 
 | | 365 A SynPspA rod after incubation with ATP | | Descriptor: | Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Non-canonical ATPase activity drives PspA membrane constriction
To Be Published
|
|
8AKX
 
 | | 305 A SynPspA rod after incubation with ATP | | Descriptor: | Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Non-canonical ATPase activity drives PspA membrane constriction
To Be Published
|
|
8AKV
 
 | | 270 A SynPspA rod after incubation with ATP | | Descriptor: | Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Non-canonical ATPase activity drives PspA membrane constriction
To Be Published
|
|
8AKW
 
 | | 280 A SynPspA rod after incubation with ATP | | Descriptor: | Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Non-canonical ATPase activity drives PspA membrane constriction
To Be Published
|
|
1FWF
 
 | |
7RNL
 
 | | Yeast CTP Synthase (Ura7) H360R Filament bound to Substrates | | Descriptor: | CTP synthase, CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Hansen, J.M, Lynch, E.M, Farrell, D.P, DiMaio, F, Quispe, J, Kollman, J.M. | | Deposit date: | 2021-07-29 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of CTP synthase filaments reveal mechanism of pH-sensitive assembly during budding yeast starvation.
Elife, 10, 2021
|
|
6YV5
 
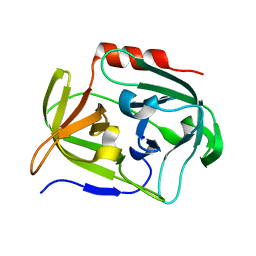 | | Crystal Structure of Serine protease SplB N3Q/S154R from Staphylococcus aureus | | Descriptor: | SODIUM ION, Serine protease | | Authors: | Rangel Pereira, M.R, Brear, P, Knyphausen, P, Jermutus, L, Hollfelder, F. | | Deposit date: | 2020-04-27 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal Structure of Serine protease SplB N3Q/S154R from Staphylococcus aureus
To Be Published
|
|
4WCX
 
 | | Crystal structure of HydG: A maturase of the [FeFe]-hydrogenase | | Descriptor: | ALANINE, Biotin and thiamin synthesis associated, FE (III) ION, ... | | Authors: | Dinis, P.C, Harmer, J.E, Driesener, R.C, Roach, P.L. | | Deposit date: | 2014-09-05 | | Release date: | 2015-02-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | X-ray crystallographic and EPR spectroscopic analysis of HydG, a maturase in [FeFe]-hydrogenase H-cluster assembly.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5VUX
 
 | | Structure of human neuronal nitric oxide synthase heme domain in complex with 7-(((4-(Dimethylamino)benzyl)amino)methyl)quinolin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[({[4-(dimethylamino)phenyl]methyl}amino)methyl]quinolin-2-amine, Nitric oxide synthase, ... | | Authors: | Huiying, L, Thomas, L.P. | | Deposit date: | 2017-05-19 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Hydrophilic, Potent, and Selective 7-Substituted 2-Aminoquinolines as Improved Human Neuronal Nitric Oxide Synthase Inhibitors.
J. Med. Chem., 60, 2017
|
|
7R9P
 
 | | Crystal structure of HPK1 in complex with compound 14 | | Descriptor: | 6-amino-2-fluoro-N,N-dimethyl-3-(4'-methylspiro[cyclopropane-1,3'-pyrrolo[2,3-b]pyridin]-5'-yl)benzamide, Hematopoietic progenitor kinase, SULFATE ION | | Authors: | Wu, P, Lehoux, I, Wang, W. | | Deposit date: | 2021-06-29 | | Release date: | 2022-01-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Discovery of Spiro-azaindoline Inhibitors of Hematopoietic Progenitor Kinase 1 (HPK1).
Acs Med.Chem.Lett., 13, 2022
|
|
7R9T
 
 | | Crystal structure of HPK1 in complex with compound 17 | | Descriptor: | 6-amino-3-[(1S,3R)-4'-chloro-3-hydroxy-1',2'-dihydrospiro[cyclopentane-1,3'-pyrrolo[2,3-b]pyridin]-5'-yl]-2-fluoro-N,N-dimethylbenzamide, Hematopoietic progenitor kinase | | Authors: | Wu, P, Lehoux, I, Wang, W. | | Deposit date: | 2021-06-29 | | Release date: | 2022-01-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Spiro-azaindoline Inhibitors of Hematopoietic Progenitor Kinase 1 (HPK1).
Acs Med.Chem.Lett., 13, 2022
|
|
7R9N
 
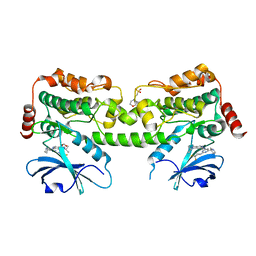 | | Crystal structure of HPK1 in complex with GNE1858 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Hematopoietic progenitor kinase, ... | | Authors: | Wu, P, Lehoux, I, Wang, W. | | Deposit date: | 2021-06-29 | | Release date: | 2022-01-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of Spiro-azaindoline Inhibitors of Hematopoietic Progenitor Kinase 1 (HPK1).
Acs Med.Chem.Lett., 13, 2022
|
|
5VUW
 
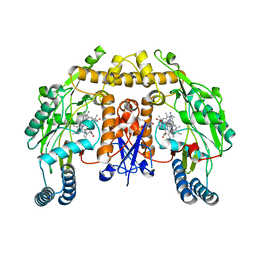 | | Structure of human neuronal nitric oxide synthase heme domain in complex with 7-(((3-(Dimethylamino)benzyl)amino)methyl)quinolin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[[[3-(dimethylamino)phenyl]methylamino]methyl]quinolin-2-amine, Nitric oxide synthase, ... | | Authors: | Huiying, L, Thomas, L.P. | | Deposit date: | 2017-05-19 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Hydrophilic, Potent, and Selective 7-Substituted 2-Aminoquinolines as Improved Human Neuronal Nitric Oxide Synthase Inhibitors.
J. Med. Chem., 60, 2017
|
|
6ZHF
 
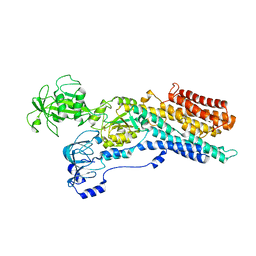 | | Calcium ATPase-1 from Listeria monocytogenes in complex with BeF | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Calcium-transporting ATPase, MAGNESIUM ION | | Authors: | Basse Hansen, S, Dyla, M, Neumann, C, Quistgaard, E.M.H, Lauwring Andersen, J, Kjaergaard, M, Nissen, P. | | Deposit date: | 2020-06-23 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | The Crystal Structure of the Ca 2+ -ATPase 1 from Listeria monocytogenes reveals a Pump Primed for Dephosphorylation.
J.Mol.Biol., 433, 2021
|
|
7RIL
 
 | | Crystal structure of hairpin polyamide Py-Im 1 bound to 5' CCTGACCAGG | | Descriptor: | 3-({3-[(3-{[4-({4-[(4-{[4-({(2R)-2-amino-4-[(1-methyl-4-{[1-methyl-4-({1-methyl-4-[(1-methyl-1H-imidazole-2-carbonyl)amino]-1H-imidazole-2-carbonyl}amino)-1H-pyrrole-2-carbonyl]amino}-1H-pyrrole-2-carbonyl)amino]butanoyl}amino)-1-methyl-1H-imidazole-2-carbonyl]amino}-1-methyl-1H-pyrrole-2-carbonyl)amino]-1-methyl-1H-pyrrole-2-carbonyl}amino)-1-methyl-1H-pyrrole-2-carbonyl]amino}propyl)(methyl)amino]propyl}carbamoyl)benzoic acid, ACETATE ION, non-template DNA, ... | | Authors: | Oh, J, Dervan, P.B, Wang, D. | | Deposit date: | 2021-07-20 | | Release date: | 2022-01-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | RNA polymerase II trapped on a molecular treadmill: Structural basis of persistent transcriptional arrest by a minor groove DNA binder.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5W6P
 
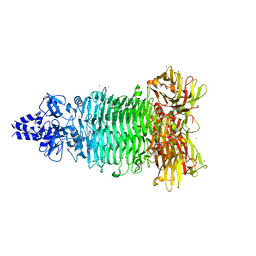 | | Crystal structure of Bacteriophage CBA120 tailspike protein 2 enzymatically active domain (TSP2dN, orf211) | | Descriptor: | 1,2-ETHANEDIOL, POTASSIUM ION, ZINC ION, ... | | Authors: | Plattner, M, Shneider, M.M, Leiman, P.G. | | Deposit date: | 2017-06-16 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.335 Å) | | Cite: | Structure and Function of the Branched Receptor-Binding Complex of Bacteriophage CBA120.
J.Mol.Biol., 431, 2019
|
|
4NP4
 
 | | Clostridium difficile toxin B CROP domain in complex with FAB domains of neutralizing antibody bezlotoxumab | | Descriptor: | Toxin B, bezlotoxumab heavy chain, bezlotoxumab light chain | | Authors: | Orth, P, Xiao, L, Hernandez, L.D, Reichert, P, Sheth, P, Beaumont, M, Murgolo, N, Ermakov, G, DiNunzio, E, Racine, F, Karczewski, J, Secore, S, Ingram, R.N, Mayhood, T, Strickland, C, Therien, A.G. | | Deposit date: | 2013-11-20 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Mechanism of Action and Epitopes of Clostridium difficile Toxin B-neutralizing Antibody Bezlotoxumab Revealed by X-ray Crystallography.
J.Biol.Chem., 289, 2014
|
|
5W67
 
 | | HLA-C*06:02 presenting VRSRR(ABA)LRL | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Mobbs, J.I, Vivian, J.P, Rossjohn, J. | | Deposit date: | 2017-06-16 | | Release date: | 2017-08-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The molecular basis for peptide repertoire selection in the human leucocyte antigen (HLA) C*06:02 molecule.
J. Biol. Chem., 292, 2017
|
|
1FWB
 
 | |
7RHY
 
 | |
