7W6Q
 
 | | Crystal structure of a PSH1 in complex with ligand J1K | | Descriptor: | 4-(2-hydroxyethylcarbamoyl)benzoic acid, PSH1 | | Authors: | Gao, J, Lara, P, Li, Z.S, Han, X, Wei, R, Liu, W.D. | | Deposit date: | 2021-12-02 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Multiple Substrate Binding Mode-Guided Engineering of a Thermophilic PET Hydrolase.
Acs Catalysis, 12, 2022
|
|
7BTS
 
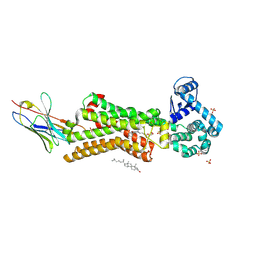 | | Structure of human beta1 adrenergic receptor bound to epinephrine and nanobody 6B9 | | Descriptor: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHOLESTEROL, ... | | Authors: | Xu, X, Kaindl, J, Clark, M, Hubner, H, Hirata, K, Sunahara, R, Gmeiner, P, Kobilka, B.K, Liu, X. | | Deposit date: | 2020-04-02 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Binding pathway determines norepinephrine selectivity for the human beta 1 AR over beta 2 AR.
Cell Res., 31, 2021
|
|
4LAZ
 
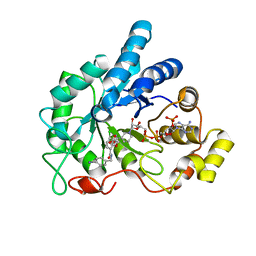 | | Crystal structure of human AR complexed with NADP+ and {5-chloro-2-[(4-iodobenzyl)carbamoyl]phenoxy}acetic acid | | Descriptor: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, {5-chloro-2-[(4-iodobenzyl)carbamoyl]phenoxy}acetic acid | | Authors: | Cousido-Siah, A, Mitschler, A, Ruiz, F.X, Fanfrlik, J, Kolar, M, Hobza, P, Podjarny, A. | | Deposit date: | 2013-06-20 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Modulation of aldose reductase inhibition by halogen bond tuning.
Acs Chem.Biol., 8, 2013
|
|
2FZF
 
 | | Hypothetical Protein Pfu-1136390-001 From Pyrococcus furiosus | | Descriptor: | hypothetical protein | | Authors: | Fu, Z.-Q, Liu, Z.-J, Lee, D, Kelley, L, Chen, L, Tempel, W, Shah, N, Horanyi, P, Lee, H.S, Habel, J, Dillard, B.D, Nguyen, D, Chang, S.-H, Zhang, H, Chang, J, Sugar, F.J, Poole, F.L, Jenney Jr, F.E, Adams, M.W.W, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-02-09 | | Release date: | 2006-02-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Hypothetical Protein Pfu-1136390-001 From Pyrococcus furiosus
To be published
|
|
5NVW
 
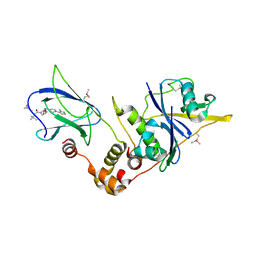 | | pVHL:EloB:EloC in complex with (2S,4R)-1-((S)-2-(cyclopropanecarboxamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 6) | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-(cyclopropylcarbonylamino)-3,3-dimethyl-butanoyl]-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Gadd, M.S, Soares, P, Galdeano, C, Ciulli, A. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Group-Based Optimization of Potent and Cell-Active Inhibitors of the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase: Structure-Activity Relationships Leading to the Chemical Probe (2S,4R)-1-((S)-2-(1-Cyanocyclopropanecarboxamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (VH298).
J. Med. Chem., 61, 2018
|
|
4IQG
 
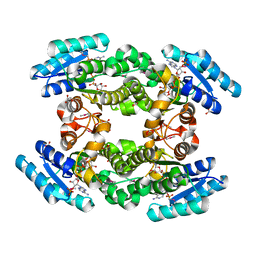 | | Crystal structure of BPRO0239 oxidoreductase from Polaromonas sp. JS666 in NADP bound form | | Descriptor: | DI(HYDROXYETHYL)ETHER, FORMIC ACID, GLYCEROL, ... | | Authors: | Niedzialkowska, E, Majorek, K.A, Porebski, P.J, Al Obaidi, N, Hammonds, J, Hillerich, B, Seidel, R, Bonanno, J.B, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-01-11 | | Release date: | 2013-01-30 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of oxidoreductase from Polaromonas sp. in NADP bound form
To be Published
|
|
1NQF
 
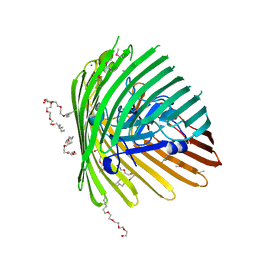 | | OUTER MEMBRANE COBALAMIN TRANSPORTER (BTUB) FROM E. COLI, METHIONINE SUBSTIUTION CONSTRUCT FOR SE-MET SAD PHASING | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, MAGNESIUM ION, Vitamin B12 receptor | | Authors: | Chimento, D.P, Mohanty, A.K, Kadner, R.J, Wiener, M.C. | | Deposit date: | 2003-01-21 | | Release date: | 2003-04-29 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Substrate-induced transmembrane signaling in the cobalamin transporter BtuB
Nat.Struct.Biol., 10, 2003
|
|
1FJE
 
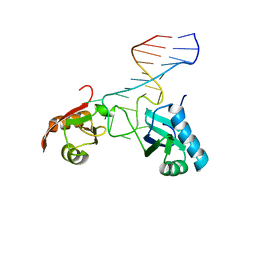 | | SOLUTION STRUCTURE OF NUCLEOLIN RBD12 IN COMPLEX WITH SNRE RNA | | Descriptor: | NUCLEOLIN RBD12, SNRE RNA | | Authors: | Allain, F.H.T, Bouvet, P, Dieckmann, T, Feigon, J. | | Deposit date: | 2000-08-08 | | Release date: | 2001-01-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular basis of sequence-specific recognition of pre-ribosomal RNA by nucleolin.
EMBO J., 19, 2000
|
|
2NC8
 
 | |
4ETY
 
 | |
2N4T
 
 | |
2G76
 
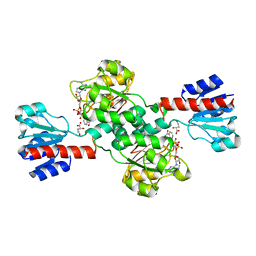 | | Crystal structure of human 3-phosphoglycerate dehydrogenase | | Descriptor: | D-3-phosphoglycerate dehydrogenase, D-MALATE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Turnbull, A.P, Salah, E, Savitsky, P, Gileadi, O, von Delft, F, Edwards, A, Arrowsmith, C, Weigelt, J, Sundstrom, M, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-02-27 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of human 3-phosphoglycerate dehydrogenase
To be Published
|
|
5O9W
 
 | | Thebaine 6-O-demethylase (T6ODM) from Papaver somniferum in complex with 2-oxoglutarate | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kluza, A, Niedzialkowska, E, Kurpiewska, K, Porebski, P.J, Borowski, T. | | Deposit date: | 2017-06-20 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of thebaine 6-O-demethylase from the morphine biosynthesis pathway.
J. Struct. Biol., 202, 2018
|
|
2YNH
 
 | | HIV-1 Reverse Transcriptase in complex with inhibitor GSK500 | | Descriptor: | 4-chloranyl-N-[[4-chloranyl-3-(3-chloranyl-5-cyano-phenoxy)-2-fluoranyl-phenyl]methyl]-2-(hydroxymethyl)-1H-imidazole-5-carboxamide, D(-)-TARTARIC ACID, P51 RT, ... | | Authors: | Chong, P, Sebahar, P, Youngman, M, Garrido, D, Zhang, H, Stewart, E.L, Nolte, R.T, Wang, L, Ferris, R.G, Edelstein, M, Weaver, K, Mathis, A, Peat, A. | | Deposit date: | 2012-10-14 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Rational Design of Potent Non-Nucleoside Inhibitors of HIV-1 Reverse Transcriptase.
J.Med.Chem., 55, 2012
|
|
3PKI
 
 | | Human SIRT6 crystal structure in complex with ADP ribose | | Descriptor: | NAD-dependent deacetylase sirtuin-6, SULFATE ION, UNKNOWN ATOM OR ION, ... | | Authors: | Pan, P.W, Dong, A, Qiu, W, Loppnau, P, Wang, J, Ravichandran, M, Bochkarev, A, Bountra, C, Weigelt, J, Arrowsmith, C.H, Min, J, Edwards, A.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-11 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure and biochemical functions of SIRT6.
J.Biol.Chem., 286, 2011
|
|
5JLY
 
 | | Structure of Peroxiredoxin-1 from Schistosoma japonicum | | Descriptor: | Thioredoxin peroxidase-1 | | Authors: | Wu, Q, Huang, F, Zeng, D, Liu, X, Zhao, J, Wang, H, Peng, Y, Li, P, Li, Y. | | Deposit date: | 2016-04-27 | | Release date: | 2017-05-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.051 Å) | | Cite: | Crystal structure of Peroxiredoxin-1 from Schistosoma japonicum
To Be Published
|
|
5JNQ
 
 | | MraY tunicamycin complex | | Descriptor: | PALMITIC ACID, Phospho-N-acetylmuramoyl-pentapeptide-transferase, Tunicamycin, ... | | Authors: | Johansson, P. | | Deposit date: | 2016-04-30 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | MraY-antibiotic complex reveals details of tunicamycin mode of action.
Nat. Chem. Biol., 13, 2017
|
|
7WOJ
 
 | | Crystal structure of HSA-Myr complex soaked with cisplatin for one week | | Descriptor: | Albumin, Cisplatin, MYRISTIC ACID | | Authors: | Chen, S.L, Yuan, C, Jiang, L.G, Luo, Z.P, Huang, M.D. | | Deposit date: | 2022-01-21 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Crystallographic analysis of interaction between cisplatin and human serum albumin: Effect of fatty acid.
Int.J.Biol.Macromol., 216, 2022
|
|
1OAU
 
 | | Fv Structure of the IgE SPE-7 in complex with DNP-Ser (immunising hapten) | | Descriptor: | 2,4-DINITROPHENOL, IMIDAZOLE, IMMUNOGLOBULIN E, ... | | Authors: | James, L.C, Roversi, P, Tawfik, D. | | Deposit date: | 2003-01-21 | | Release date: | 2004-01-15 | | Last modified: | 2013-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Antibody Multispecificity Mediated by Conformational Diversity
Science, 299, 2003
|
|
4ENW
 
 | | Structure of the S234N variant of E. coli KatE | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, Catalase HPII | | Authors: | Loewen, P.C, Jha, V. | | Deposit date: | 2012-04-13 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Influence of main channel structure on H(2)O(2) access to the heme cavity of catalase KatE of Escherichia coli.
Arch.Biochem.Biophys., 526, 2012
|
|
2YLH
 
 | |
4E6S
 
 | | Crystal structure of the SCAN domain from mouse Zfp206 | | Descriptor: | Zinc finger and SCAN domain-containing protein 10 | | Authors: | Liang, Y, Choo, S.H, Rossbach, M, Baburajendran, N, Palasingam, P, Kolatkar, P.R. | | Deposit date: | 2012-03-15 | | Release date: | 2012-05-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal optimization and preliminary diffraction data analysis of the SCAN domain of Zfp206.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
2YNG
 
 | | HIV-1 Reverse Transcriptase in complex with inhibitor GSK560 | | Descriptor: | 2-azanyl-N-[[4-bromanyl-3-(3-chloranyl-5-cyano-phenoxy)-2-fluoranyl-phenyl]methyl]-4-chloranyl-1H-imidazole-5-carboxamide, MAGNESIUM ION, P51 RT, ... | | Authors: | Chong, P, Sebahar, P, Youngman, M, Garrido, D, Zhang, H, Stewart, E.L, Nolte, R.T, Wang, L, Ferris, R.G, Edelstein, M, Weaver, K, Mathis, A, Peat, A. | | Deposit date: | 2012-10-14 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Rational Design of Potent Non-Nucleoside Inhibitors of HIV-1 Reverse Transcriptase.
J.Med.Chem., 55, 2012
|
|
4JDT
 
 | | Crystal structure of chimeric germ-line precursor of NIH45-46 Fab in complex with gp120 of 93TH057 HIV-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab heavy chain, Fab light chain, ... | | Authors: | Scharf, L, Diskin, R, Bjorkman, P.J. | | Deposit date: | 2013-02-25 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.26 Å) | | Cite: | Structural basis for HIV-1 gp120 recognition by a germ-line version of a broadly neutralizing antibody.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JEC
 
 | | Joint neutron and X-ray structure of per-deuterated HIV-1 protease in complex with clinical inhibitor amprenavir | | Descriptor: | CHLORIDE ION, HIV-1 protease, {3-[(4-AMINO-BENZENESULFONYL)-ISOBUTYL-AMINO]-1-BENZYL-2-HYDROXY-PROPYL}-CARBAMIC ACID TETRAHYDRO-FURAN-3-YL ESTER | | Authors: | Kovalevsky, A.Y, Weber, I.T, Langan, P. | | Deposit date: | 2013-02-26 | | Release date: | 2013-07-24 | | Last modified: | 2024-02-28 | | Method: | NEUTRON DIFFRACTION (2.01 Å), X-RAY DIFFRACTION | | Cite: | Joint X-ray/Neutron Crystallographic Study of HIV-1 Protease with Clinical Inhibitor Amprenavir: Insights for Drug Design.
J.Med.Chem., 56, 2013
|
|
