4CV2
 
 | | Crystal structure of E. coli FabI in complex with NADH and CG400549 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 1-(3-amino-2-methylbenzyl)-4-[2-(thiophen-2-yl)ethoxy]pyridin-2(1H)-one, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH] | | Authors: | Tareilus, M, Schiebel, J, Chang, A, Shah, S, Tonge, P.J, Kisker, C. | | Deposit date: | 2014-03-22 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rational Design of Broad Spectrum Antibacterial Activity Based on a Clinically Relevant Enoyl-Acyl Carrier Protein (Acp) Reductase Inhibitor.
J.Biol.Chem., 289, 2014
|
|
5I22
 
 | | Amphiphysin SH3 in complex with Chikungunya virus nsP3 peptide | | Descriptor: | CHIKV nsP3 peptide, Myc box-dependent-interacting protein 1 | | Authors: | Tossavainen, H, Aitio, O, Hellman, M, Saksela, K, Permi, P. | | Deposit date: | 2016-02-04 | | Release date: | 2016-06-15 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of the High Affinity Interaction between the Alphavirus Nonstructural Protein-3 (nsP3) and the SH3 Domain of Amphiphysin-2.
J.Biol.Chem., 291, 2016
|
|
2J95
 
 | | CRYSTAL STRUCTURE OF A HUMAN FACTOR XA INHIBITOR COMPLEX | | Descriptor: | 5'-CHLORO-N-{(3S)-1-[(1S)-1-METHYL-2-MORPHOLIN-4-YL-2-OXOETHYL]-2-OXOPYRROLIDIN-3-YL}-2,2'-BITHIOPHENE-5-SULFONAMIDE, ACTIVATED FACTOR XA HEAVY CHAIN, ACTIVATED FACTOR XA LIGHT CHAIN, ... | | Authors: | Chan, C, Borthwick, A.D, Brown, D, Campbell, M, Chaudry, L, Chung, C.W, Convery, M.A, Hamblin, J.N, Johnstone, L, Kelly, H.A, Kleanthous, S, Burns-Kurtis, C.L, Patikis, A, Patel, C, Pateman, A.J, Senger, S, Shah, G.P, Toomey, J.R, Watson, N.S, Weston, H.E, Whitworth, C, Young, R.J, Zhou, P. | | Deposit date: | 2006-11-02 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Factor Xa Inhibitors: S1 Binding Interactions of a Series of N-{(3S)-1-[(1S)-1-Methyl-2-Morpholin-4-Yl-2-Oxoethyl]-2-Oxopyrrolidin-3-Yl}Sulfonamides.
J.Med.Chem., 50, 2007
|
|
2IEN
 
 | | Crystal structure analysis of HIV-1 protease with a potent non-peptide inhibitor (UIC-94017) | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Manna, D, Hussain, A.K, Leshchenko, S, Ghosh, A.K, Louis, J.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2006-09-19 | | Release date: | 2006-10-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High Resolution Crystal Structures of HIV-1 Protease with a Potent Non-Peptide Inhibitor (Uic-94017) Active Against Multi-Drug-Resistant Clinical Strains.
J.Mol.Biol., 338, 2004
|
|
4PFP
 
 | | Myosin VI motor domain in the Pi release state (with Pi) space group P21 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Isabet, T, Benisty, H, Llinas, P, Sweeney, H.L, Houdusse, A. | | Deposit date: | 2014-04-30 | | Release date: | 2015-04-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | How actin initiates the motor activity of Myosin.
Dev.Cell, 33, 2015
|
|
4PHF
 
 | | Crystal structure of Ypt7 covalently modified with GDP | | Descriptor: | GTP-binding protein YPT7, MAGNESIUM ION, N-[3-(propanoylamino)propyl]guanosine 5'-(trihydrogen diphosphate), ... | | Authors: | Vieweg, S, Wiegandt, D, Hofmann, F, Koch, D, Wu, Y, Itzen, A, Mueller, M.P, Goody, R.S. | | Deposit date: | 2014-05-06 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Locking GTPases covalently in their functional states.
Nat Commun, 6, 2015
|
|
1IVT
 
 | | NMR structures of the C-terminal globular domain of human lamin A/C | | Descriptor: | Lamin A/C | | Authors: | Krimm, I, Ostlund, C, Gilquin, B, Couprie, J, Hossenlopp, P, Mornon, J.P, Bonn, G, Courvalin, J.C, Worman, H.J, Zinn-Justin, S. | | Deposit date: | 2002-03-29 | | Release date: | 2002-08-21 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The Ig-like structure of the C-terminal domain of lamin A/C, mutated in muscular dystrophies, cardiomyopathy, and partial lipodystrophy.
Structure, 10, 2002
|
|
1PAE
 
 | | nucleoside diphosphate kinase | | Descriptor: | Nucleoside diphosphate kinase, cytosolic, SELENIUM ATOM | | Authors: | Strub, M.-P, Hoh, F, Sanchez, J.-F, Strub, J.M, Bock, A, Aumelas, A, Dumas, C. | | Deposit date: | 2003-05-14 | | Release date: | 2003-11-11 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Selenomethionine and Selenocysteine Double Labeling Strategy for Crystallographic Phasing
Structure, 11, 2003
|
|
4PJJ
 
 | | MYOSIN VI (MD-INSERT2-CAM, DELTA-INSERT1) post-rigor state - long soaking with PO4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, Calmodulin, ... | | Authors: | Isabet, T, Benisty, H, Llinas, P, Sweeney, H.L, Houdusse, A. | | Deposit date: | 2014-05-12 | | Release date: | 2015-04-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | How actin initiates the motor activity of Myosin.
Dev.Cell, 33, 2015
|
|
4PJN
 
 | | Myosin VI motor domain in the Pi release state, space group P212121 - shortly soaked with PO4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Isabet, T, Benisty, H, Llinas, P, Sweeney, H.L, Houdusse, A. | | Deposit date: | 2014-05-12 | | Release date: | 2015-04-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | How actin initiates the motor activity of Myosin.
Dev.Cell, 33, 2015
|
|
2J1O
 
 | | Geranylgeranyl diphosphate synthase from Sinapis alba | | Descriptor: | GERANYLGERANYL PYROPHOSPHATE SYNTHETASE, GLYCEROL | | Authors: | Kloer, D.P, Welsch, R, Beyer, P, Schulz, G.E. | | Deposit date: | 2006-08-15 | | Release date: | 2007-01-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Reaction Geometry of Geranylgeranyl Diphosphate Synthase from Sinapis Alba.
Biochemistry, 45, 2006
|
|
1SFF
 
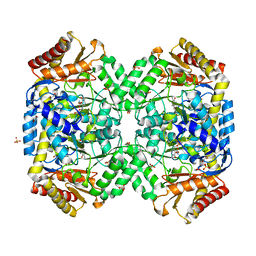 | | Structure of gamma-aminobutyrate aminotransferase complex with aminooxyacetate | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-ACETYLYAMINO-PYRIDOXAL-5'-PHOSPHATE, 4-aminobutyrate aminotransferase, ... | | Authors: | Liu, W, Peterson, P.E, Carter, R.J, Zhou, X, Langston, J.A, Fisher, A.J, Toney, M.D. | | Deposit date: | 2004-02-19 | | Release date: | 2004-09-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of unbound and aminooxyacetate-bound Escherichia coli gamma-aminobutyrate aminotransferase.
Biochemistry, 43, 2004
|
|
4D4L
 
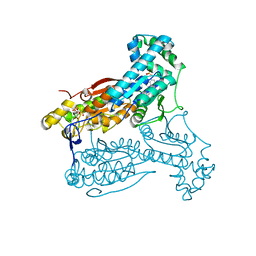 | | human PFKFB3 in complex with a pyrrolopyrimidone compound | | Descriptor: | 5-(4-chlorophenyl)-7-phenyl-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, 6-O-phosphono-beta-D-fructofuranose, 6-PHOSPHOFRUCTO-2-KINASE/FRUCTOSE-2,6-BISPHOSPHATASE 3, ... | | Authors: | Stgallay, S.A, Bennett, N, Critchlow, S, Davies, G, Debreczeni, J.E, Evans, N, Holdgate, G, Jones, N.P, Leach, L, Maman, S, McLoughlin, S, Preston, M, Rigoreau, L, Thomas, A, Walker, G, Walsch, J, Ward, R.A, Wheatley, E, Winter, J. | | Deposit date: | 2014-10-29 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Identifying a Novel Series of Pfkfb3 Inhibitors as a Metabolic Approach to Treating Cancer from Hts, Biophysical and Biochemical Methods
To be Published
|
|
2IQC
 
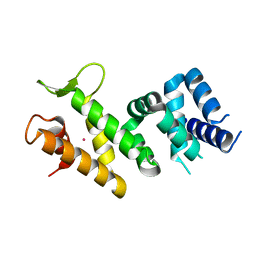 | | Crystal structure of Human FancF Protein that Functions in the Assembly of a DNA Damage Signaling Complex | | Descriptor: | Fanconi anemia group F protein, MERCURY (II) ION | | Authors: | Kowal, P, Gurtan, A.M, Stuckert, P, Lehmann, C, D'Andrea, A, Ellenberger, T.E. | | Deposit date: | 2006-10-13 | | Release date: | 2006-11-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural determinants of human FANCF protein that function in the assembly of a DNA damage signaling complex.
J.Biol.Chem., 282, 2007
|
|
3HT8
 
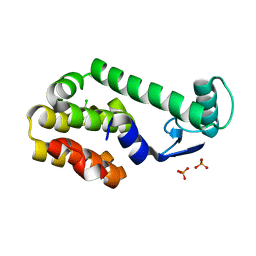 | | 5-chloro-2-methylphenol in complex with T4 lysozyme L99A/M102Q | | Descriptor: | 5-chloro-2-methylphenol, Lysozyme, PHOSPHATE ION | | Authors: | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | Deposit date: | 2009-06-11 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
4M5A
 
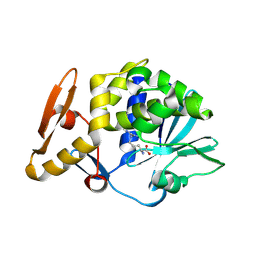 | | Crystal structure of the complex of Ribosome inactivating protein from Momordica balsamina inhibited by asymmetric dimethyl arginine at 1.70 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NG,NG-DIMETHYL-L-ARGININE, rRNA N-glycosidase | | Authors: | Yamini, S, Pandey, S, Kushwaha, G.S, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-08-08 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the complex of Ribosome inactivating protein from Momordica balsamina inhibited by asymmetric dimethyl arginine at 1.70 A resolution
To be Published
|
|
3HTG
 
 | | 2-ethoxy-3,4-dihydro-2h-pyran in complex with T4 lysozyme L99A/M102Q | | Descriptor: | (2S)-2-ethoxy-3,4-dihydro-2H-pyran, BETA-MERCAPTOETHANOL, Lysozyme, ... | | Authors: | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | Deposit date: | 2009-06-11 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
1P6A
 
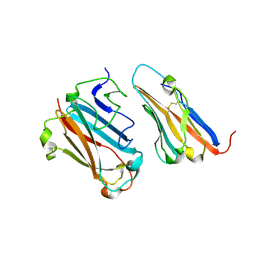 | | STRUCTURAL BASIS FOR VARIATION IN ADENOVIRUS AFFINITY FOR THE CELLULAR RECEPTOR CAR (S489Y MUTANT) | | Descriptor: | Coxsackievirus and adenovirus receptor, Fiber protein | | Authors: | Howitt, J, Bewley, M.C, Graziano, V, Flanagan, J.M, Freimuth, P. | | Deposit date: | 2003-04-29 | | Release date: | 2004-05-11 | | Last modified: | 2018-08-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for variation in adenovirus affinity for the cellular coxsackievirus and adenovirus receptor.
J.Biol.Chem., 278, 2003
|
|
3HTZ
 
 | |
3HUA
 
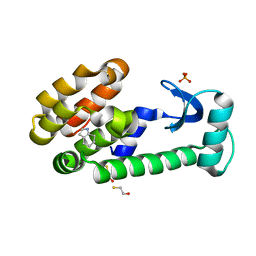 | | 4,5,6,7-tetrahydroindole in complex with T4 lysozyme L99A/M102Q | | Descriptor: | 4,5,6,7-tetrahydro-1H-indole, BETA-MERCAPTOETHANOL, Lysozyme, ... | | Authors: | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | Deposit date: | 2009-06-13 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
3HU8
 
 | | 2-ethoxyphenol in complex with T4 lysozyme L99A/M102Q | | Descriptor: | 2-ethoxyphenol, Lysozyme, PHOSPHATE ION | | Authors: | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | Deposit date: | 2009-06-13 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
7AV9
 
 | | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group C2 | | Descriptor: | 1,2-ETHANEDIOL, PH-interacting protein | | Authors: | Krojer, T, Talon, R, Fairhead, M, Szykowska, A, Burgess-Brown, N.A, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F. | | Deposit date: | 2020-11-04 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group C2
To Be Published
|
|
5I2A
 
 | | 1,2-propanediol Dehydration in Roseburia inulinivorans; Structural Basis for Substrate and Enantiomer Selectivity | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Diol-dehydratase | | Authors: | LaMattina, J.W, Reitzer, P, Kapoor, S, Galzerani, F, Koch, D.J, Gouvea, I.E, Lanzilotta, W.N. | | Deposit date: | 2016-02-08 | | Release date: | 2016-06-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 1,2-Propanediol Dehydration in Roseburia inulinivorans: STRUCTURAL BASIS FOR SUBSTRATE AND ENANTIOMER SELECTIVITY.
J.Biol.Chem., 291, 2016
|
|
7JZQ
 
 | |
1OMI
 
 | | Crystal structure of PrfA,the transcriptional regulator in Listeria monocytogenes | | Descriptor: | GLYCEROL, Listeriolysin regulatory protein | | Authors: | Thirumuruhan, R, Rajashankar, K, Fedorov, A.A, Dodatko, T, Chance, M.R, Cossart, P, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-02-25 | | Release date: | 2003-03-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of PrfA, the transcriptional regulator in Listeria monocytogenes
To be Published
|
|
