1D6Z
 
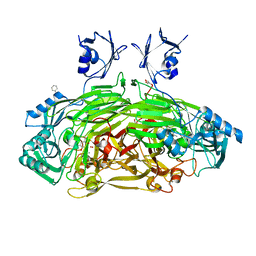 | | CRYSTAL STRUCTURE OF THE AEROBICALLY FREEZE TRAPPED RATE-DETERMINING CATALYTIC INTERMEDIATE OF E. COLI COPPER-CONTAINING AMINE OXIDASE. | | Descriptor: | 2-PHENYLETHYLAMINE, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Wilmot, C.M, Hajdu, J, McPherson, M.J, Knowles, P.F, Phillips, S.E.V. | | Deposit date: | 1999-10-16 | | Release date: | 2000-02-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Visualization of dioxygen bound to copper during enzyme catalysis.
Science, 286, 1999
|
|
2PXE
 
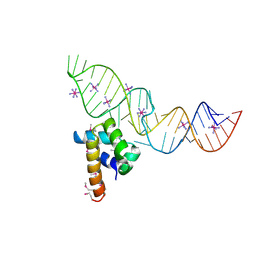 | | Variant 4 of Ribonucleoprotein Core of the E. Coli Signal Recognition Particle | | Descriptor: | 4.5 S RNA, COBALT HEXAMMINE(III), Signal recognition particle protein | | Authors: | Keel, A.Y, Rambo, R.P, Batey, R.T, Kieft, J.S. | | Deposit date: | 2007-05-14 | | Release date: | 2007-08-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A General Strategy to Solve the Phase Problem in RNA Crystallography.
Structure, 15, 2007
|
|
2A08
 
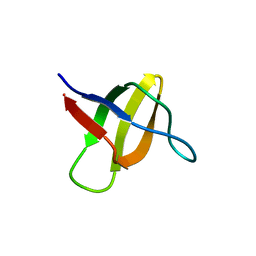 | | Structure of the yeast YHH6 SH3 domain | | Descriptor: | Hypothetical 41.8 kDa protein in SPO13-ARG4 intergenic region | | Authors: | Kursula, P, Kursula, I, Song, Y.H, Lehmann, F, Zou, P, Wilmanns, M. | | Deposit date: | 2005-06-16 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | 3-D proteome of yeast SH3 domains
To be Published
|
|
4B95
 
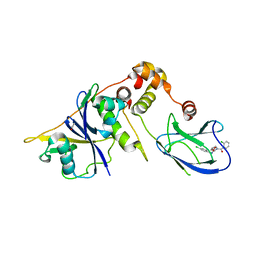 | | pVHL-EloB-EloB-EloC complex_(2S,4R)-1-(2-chlorophenyl)carbonyl-N-[(4-chlorophenyl)methyl]-4-oxidanyl-pyrrolidine-2-carboxamide bound | | Descriptor: | (2S,4R)-1-(2-chlorophenyl)carbonyl-N-[(4-chlorophenyl)methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, ACETATE ION, TRANSCRIPTION ELONGATION FACTOR B POLYPEPTIDE 1, ... | | Authors: | Buckley, D.L, Gustafson, J.L, VanMolle, I, Roth, A.G, SeopTae, H, Gareiss, P.C, Jorgensen, W.L, Ciulli, A, Crews, C.M. | | Deposit date: | 2012-08-31 | | Release date: | 2012-10-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Small-Molecule Inhibitors of the Interaction between the E3 Ligase Vhl and Hif1Alpha
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4D1D
 
 | | STRUCTURE OF MHP1, A NUCLEOBASE-CATION-SYMPORT-1 FAMILY TRANSPORTER with the inhibitor 5-(2-naphthylmethyl)-L-hydantoin. | | Descriptor: | 5-(2-NAPHTHYLMETHYL)-D-HYDANTOIN, 5-(2-NAPHTHYLMETHYL)-L-HYDANTOIN, HYDANTOIN TRANSPORT PROTEIN, ... | | Authors: | Weyand, S, Brueckner, F, Geng, T, Drew, D, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Molecular Mechanism of Ligand Recognition by Membrane Transport Protein, Mhp1.
Embo J., 33, 2014
|
|
1Z3Q
 
 | | Resolution of the structure of the allergenic and antifungal banana fruit thaumatin-like protein at 1.7A | | Descriptor: | 1,2-ETHANEDIOL, Thaumatin-like Protein | | Authors: | Leone, P, Menu-Bouaouiche, L, Peumans, W.J, Barre, A, Payan, F, Roussel, A, Van Damme, E.J.M, Rouge, P. | | Deposit date: | 2005-03-14 | | Release date: | 2006-01-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Resolution of the structure of the allergenic and antifungal banana fruit thaumatin-like protein at 1.7-A
Biochimie, 88, 2006
|
|
1SRN
 
 | |
4B6D
 
 | | Structure of the atypical C1 domain of MgcRacGAP | | Descriptor: | GLYCEROL, RAC GTPASE-ACTIVATING PROTEIN 1, ZINC ION | | Authors: | Pye, V.E, Lekomtsev, S, Petronczki, M, Cherepanov, P. | | Deposit date: | 2012-08-09 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Centralspindlin Links the Mitotic Spindle to the Plasma Membrane During Cytokinesis.
Nature, 492, 2012
|
|
1Z63
 
 | | Sulfolobus solfataricus SWI2/SNF2 ATPase core in complex with dsDNA | | Descriptor: | 5'-D(*AP*AP*AP*AP*AP*A*AP*TP*TP*GP*CP*CP*GP*AP*AP*GP*AP*CP*GP*AP*AP*AP*AP*AP*A)-3', 5'-D(*TP*TP*TP*TP*TP*TP*TP*CP*GP*TP*CP*TP*TP*CP*GP*GP*CP*AP*AP*TP*TP*TP*TP*TP*T)-3', Helicase of the snf2/rad54 family | | Authors: | Duerr, H, Koerner, C, Mueller, M, Hickmann, V, Hopfner, K.P. | | Deposit date: | 2005-03-21 | | Release date: | 2005-05-17 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with DNA.
Cell(Cambridge,Mass.), 121, 2005
|
|
1Z75
 
 | | Crystal Structure of ArnA dehydrogenase (decarboxylase) domain, R619M mutant | | Descriptor: | GLYCEROL, SULFATE ION, protein ArnA | | Authors: | Gatzeva-Topalova, P.Z, May, A.P, Sousa, M.C. | | Deposit date: | 2005-03-24 | | Release date: | 2005-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and Mechanism of ArnA: Conformational Change Implies Ordered Dehydrogenase Mechanism in Key Enzyme for Polymyxin Resistance
Structure, 13, 2005
|
|
4B4O
 
 | | Crystal Structure of human epimerase family protein SDR39U1 (isoform2) with NADPH | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, EPIMERASE FAMILY PROTEIN SDR39U1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Vollmar, M, Muniz, J.R.C, Shafqat, N, Picaud, S, Krojer, T, Chaikuad, A, Pike, A.C.W, Yue, W.W, Filippakopoulos, P, Kavanagh, K.L, von Delft, F, Weigelt, J, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U. | | Deposit date: | 2012-07-31 | | Release date: | 2012-08-29 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Human Epimerase Family Protein Sdr39U1 (Isoform2) with Nadph
To be Published
|
|
2XKQ
 
 | | Crystal structure of Streptococcus suis Dpr with manganese | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Haikarainen, T, Thanassoulas, A, Stavros, P, Nounesis, G, Haataja, S, Papageorgiou, A.C. | | Deposit date: | 2010-07-12 | | Release date: | 2010-11-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Thermodynamic Characterization of Metal Ion Binding in Streptococcus Suis Dpr.
J.Mol.Biol., 405, 2011
|
|
4D0W
 
 | | Pyrrole-3-carboxamides as potent and selective JAK2 inhibitors | | Descriptor: | 5-(2-aminopyrimidin-4-yl)-2-(5-chloro-2-methylphenyl)-1H-pyrrole-3-carboxamide, GLYCEROL, TYROSINE-PROTEIN KINASE JAK2 | | Authors: | Bertrand, J, Canevari, G, Fasolini, M, Brasca, M.G, Nesi, M, Avanzi, N, Ballinari, D, Bandiera, T, Bindi, S, Carenzi, D, Casero, D, Ceriani, L, Ciomei, M, Cirla, A, Colombo, M, Cribioli, S, Cristiani, C, Della Vedova, F, Fachin, G, Felder, E.R, Galvani, A, Isacchi, A, Mirizzi, D, Motto, I, Panzeri, A, Pesenti, E, Vianello, P, Gnocchi, P, Donati, D. | | Deposit date: | 2014-04-30 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Pyrrole-3-Carboxamides as Potent and Selective Jak2 Inhibitors.
Bioorg.Med.Chem., 22, 2014
|
|
4D1S
 
 | | Pyrrole-3-carboxamides as potent and selective JAK2 inhibitors | | Descriptor: | 2-(5-chloro-2-methylphenyl)-1-methyl-5-(2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)-1H-pyrrole-3-carboxamide, TYROSINE-PROTEIN KINASE JAK2 | | Authors: | Bertrand, J, Canevari, G, Fasolini, M, Brasca, M.G, Nesi, M, Avanzi, N, Ballinari, D, Bandiera, T, Bindi, S, Carenzi, D, Casero, D, Ceriani, L, Ciomei, M, Cirla, A, Colombo, M, Cribioli, S, Cristiani, C, Della Vedova, F, Fachin, G, Felder, E.R, Galvani, A, Isacchi, A, Mirizzi, D, Motto, I, Panzeri, A, Pesenti, E, Vianello, P, Gnocchi, P, Donati, D. | | Deposit date: | 2014-05-05 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Pyrrole-3-Carboxamides as Potent and Selective Jak2 Inhibitors.
Bioorg.Med.Chem., 22, 2014
|
|
1Z8V
 
 | | The Structure of d(GGCCAATTGG) Complexed with Netropsin | | Descriptor: | (5'-D(*GP*GP*CP*CP*AP*AP*TP*TP*GP*G)-3'), NETROPSIN | | Authors: | Van Hecke, K, Nam, P.C, Nguyen, M.T, Van Meervelt, L. | | Deposit date: | 2005-03-31 | | Release date: | 2006-03-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Netropsin interactions in the minor groove of d(GGCCAATTGG) studied by a combination of resolution enhancement and ab initio calculations.
Febs J., 272, 2005
|
|
1ZAG
 
 | | HUMAN ZINC-ALPHA-2-GLYCOPROTEIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chirino, A.J, Sanchez, L.M, Bjorkman, P.J. | | Deposit date: | 1999-02-02 | | Release date: | 1999-03-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human ZAG, a fat-depleting factor related to MHC molecules.
Science, 283, 1999
|
|
3T9Z
 
 | | A. fulgidus GlnK3, ligand-free | | Descriptor: | CITRATE ANION, Nitrogen regulatory protein P-II (GlnB-3) | | Authors: | Maier, S, Schleberger, P, Lue, W, Wacker, T, Pflueger, T, Litz, C, Andrade, S.L.A. | | Deposit date: | 2011-08-03 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Mechanism of disruption of the Amt-GlnK complex by P(II)-mediated sensing of 2-oxoglutarate.
Plos One, 6, 2011
|
|
1ZBC
 
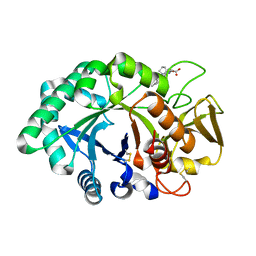 | | Crystal Structure of the porcine signalling protein liganded with the peptide Trp-Pro-Trp (WPW) at 2.3 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3 mer peptide, signal processing protein | | Authors: | Srivastava, D.B, Kaur, P, Kumar, J, Somvanshi, R.K, Sharma, S, Dey, S, Singh, T.P. | | Deposit date: | 2005-04-08 | | Release date: | 2005-04-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal Structure of the porcine signalling protein liganded with the peptide Trp-Pro-Trp (WPW) at 2.3 A resolution
To be Published
|
|
2GLK
 
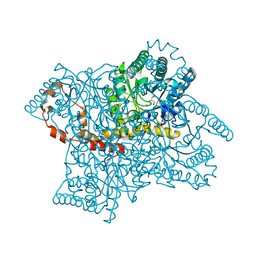 | | High-resolution study of D-Xylose isomerase, 0.94A resolution. | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Xylose isomerase | | Authors: | Katz, A.K, Carrell, H.L, Hanson, B.L, Harp, J.M, Glusker, J.P, Bunick, G.J. | | Deposit date: | 2006-04-05 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Locating active-site hydrogen atoms in D-xylose isomerase: Time-of-flight neutron diffraction.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
3PFV
 
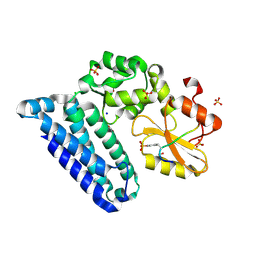 | | Crystal structure of Cbl-b TKB domain in complex with EGFR pY1069 peptide | | Descriptor: | 1,2-ETHANEDIOL, 11-meric peptide from Epidermal growth factor receptor, CHLORIDE ION, ... | | Authors: | Chaikuad, A, Guo, K, Cooper, C.D.O, Ayinampudi, V, Krojer, T, Muniz, J.R.C, Vollmar, M, Canning, P, Gileadi, O, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-10-29 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of Cbl-b TKB domain in complex with EGFR pY1069 peptide
To be Published
|
|
4D0X
 
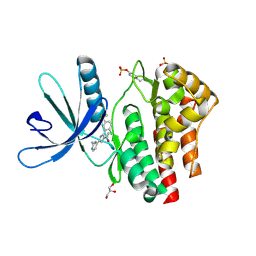 | | Pyrrole-3-carboxamides as potent and selective JAK2 inhibitors | | Descriptor: | 5-(2-aminopyrimidin-4-yl)-2-[2-chloro-5-(trifluoromethyl)phenyl]-1H-pyrrole-3-carboxamide, GLYCEROL, TYROSINE-PROTEIN KINASE JAK2 | | Authors: | Canevari, G, Fasolini, M, Bertrand, J, Brasca, M.G, Nesi, M, Avanzi, N, Ballinari, D, Bandiera, T, Bindi, S, Carenzi, D, Casero, D, Ceriani, L, Ciomei, M, Cirla, A, Colombo, M, Cribioli, S, Cristiani, C, Della Vedova, F, Fachin, G, Felder, E.R, Galvani, A, Isacchi, A, Mirizzi, D, Motto, I, Panzeri, A, Pesenti, E, Vianello, P, Gnocchi, P, Donati, D. | | Deposit date: | 2014-04-30 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Pyrrole-3-Carboxamides as Potent and Selective Jak2 Inhibitors.
Bioorg.Med.Chem., 22, 2014
|
|
4D1A
 
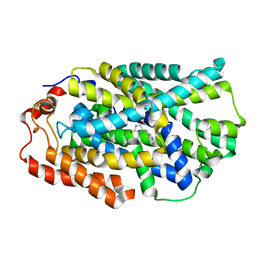 | | STRUCTURE OF MHP1, A NUCLEOBASE-CATION-SYMPORT-1 FAMILY TRANSPORTER, IN A CLOSED CONFORMATION WITH INDOLYLMETHYL-HYDANTOIN | | Descriptor: | (5S)-5-(1H-indol-3-ylmethyl)imidazolidine-2,4-dione, HYDANTOIN TRANSPORT PROTEIN, SODIUM ION | | Authors: | Weyand, S, Brueckner, F, Geng, T, Drew, D, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Molecular Mechanism of Ligand Recognition by Membrane Transport Protein, Mhp1.
Embo J., 33, 2014
|
|
2XLL
 
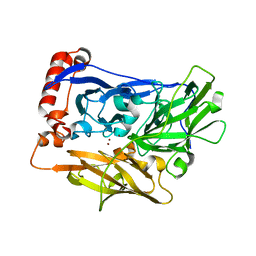 | | The crystal structure of bilirubin oxidase from Myrothecium verrucaria | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIRUBIN OXIDASE, COPPER (II) ION | | Authors: | McNamara, T.P, Lowe, E.D, Cracknell, J.A, Blanford, C.F. | | Deposit date: | 2010-07-21 | | Release date: | 2011-04-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.305 Å) | | Cite: | Bilirubin Oxidase from Myrothecium Verrucaria: X- Ray Determination of the Complete Crystal Structure and a Rational Surface Modification for Enhanced Electrocatalytic O(2) Reduction.
Dalton Trans, 40, 2011
|
|
1ZFA
 
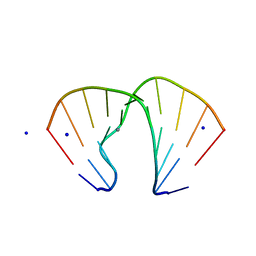 | | GGA Duplex A-DNA | | Descriptor: | 5'-D(*CP*CP*TP*CP*CP*GP*GP*AP*GP*G)-3', CALCIUM ION, SODIUM ION | | Authors: | Hays, F.A, Teegarden, A.T, Jones, Z.J.R, Harms, M, Raup, D, Watson, J, Cavaliere, E, Ho, P.S. | | Deposit date: | 2005-04-20 | | Release date: | 2005-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | How sequence defines structure: a crystallographic map of DNA structure and conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
4D2O
 
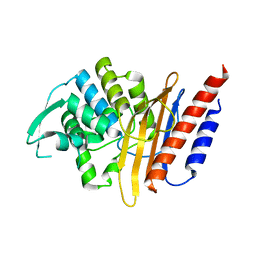 | | Crystal structure of the class A extended-spectrum beta-lactamase PER- 2 | | Descriptor: | PER-2 BETA-LACTAMASE | | Authors: | Power, P, Herman, R, Ruggiero, M, Kerff, F, Galleni, M, Gutkind, G, Charlier, P, Sauvage, E. | | Deposit date: | 2014-05-12 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Extended-Spectrum Beta-Lactamase Per- 2 and Insights Into the Role of Specific Residues in the Interaction with Beta-Lactams and Beta-Lactamase Inhibitors.
Antimicrob.Agents Chemother., 58, 2014
|
|
