5KR0
 
 | | Protease E35D-APV | | Descriptor: | GLYCEROL, Protease E35D-APV, {3-[(4-AMINO-BENZENESULFONYL)-ISOBUTYL-AMINO]-1-BENZYL-2-HYDROXY-PROPYL}-CARBAMIC ACID TETRAHYDRO-FURAN-3-YL ESTER | | Authors: | Liu, Z, Poole, K.M, Mahon, B.P, McKenna, R, Fanucci, G.E. | | Deposit date: | 2016-07-06 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Effects of Hinge-region Natural Polymorphisms on Human Immunodeficiency Virus-Type 1 Protease Structure, Dynamics, and Drug Pressure Evolution.
J.Biol.Chem., 291, 2016
|
|
7QGJ
 
 | | Apo structure of BIR2 Domain of BIRC2 | | Descriptor: | 1,2-ETHANEDIOL, Baculoviral IAP repeat-containing protein 2, ZINC ION | | Authors: | Kraemer, A, Farges, F, Schwalm, M.P, Saxena, K, Preuss, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-12-08 | | Release date: | 2022-02-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Apo structure BIR2 Domain of BIRC2
To Be Published
|
|
1S9N
 
 | | Solution structure of the nitrous acid (G)-(G) cross-linked DNA dodecamer duplex GCATCC(G)GATGC | | Descriptor: | 5'-D(*GP*CP*AP*TP*CP*CP*GP*GP*AP*TP*GP*C)-3' | | Authors: | Edfeldt, N.B.F, Harwood, E.A, Sigurdsson, S.T, Hopkins, P.B, Reid, B.R. | | Deposit date: | 2004-02-05 | | Release date: | 2005-06-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a nitrous acid induced DNA interstrand cross-link
Nucleic Acids Res., 32, 2004
|
|
4DVE
 
 | | Crystal structure at 2.1 A of the S-component for biotin from an ECF-type ABC transporter | | Descriptor: | BIOTIN, Biotin transporter BioY, nonyl beta-D-glucopyranoside | | Authors: | Berntsson, R.P.-A, ter Beek, J, Majsnerowska, M, Duurkens, R, Puri, P, Poolman, B, Slotboom, D.J. | | Deposit date: | 2012-02-23 | | Release date: | 2012-08-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural divergence of paralogous S components from ECF-type ABC transporters.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5BN3
 
 | | Structure of a unique ATP synthase NeqA-NeqB in complex with ADP from Nanoarcheaum equitans | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Mohanty, S, Jobichen, C, Chichili, V.P.R, Sivaraman, J. | | Deposit date: | 2015-05-25 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for a Unique ATP Synthase Core Complex from Nanoarcheaum equitans
J.Biol.Chem., 290, 2015
|
|
1AZG
 
 | | NMR STUDY OF THE SH3 DOMAIN FROM FYN PROTO-ONCOGENE TYROSINE KINASE KINASE COMPLEXED WITH THE SYNTHETIC PEPTIDE P2L CORRESPONDING TO RESIDUES 91-104 OF THE P85 SUBUNIT OF PI3-KINASE, MINIMIZED AVERAGE (PROBMAP) STRUCTURE | | Descriptor: | FYN, PRO-PRO-ARG-PRO-LEU-PRO-VAL-ALA-PRO-GLY-SER-SER-LYS-THR | | Authors: | Renzoni, D.A, Pugh, D.J.R, Siligardi, G, Das, P, Morton, C.J, Rossi, C, Waterfield, M.D, Campbell, I.D, Ladbury, J.E. | | Deposit date: | 1997-11-18 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural and thermodynamic characterization of the interaction of the SH3 domain from Fyn with the proline-rich binding site on the p85 subunit of PI3-kinase.
Biochemistry, 35, 1996
|
|
5TIN
 
 | | Crystal Structure of Human Glycine Receptor alpha-3 Mutant N38Q Bound to AM-3607 | | Descriptor: | (3S,3aS,9bS)-2-[(2H-1,3-benzodioxol-5-yl)sulfonyl]-3,5-dimethyl-1,2,3,3a,5,9b-hexahydro-4H-pyrrolo[3,4-c][1,6]naphthyridin-4-one, CHLORIDE ION, GLYCINE, ... | | Authors: | Shaffer, P.L, Huang, X, Chen, H. | | Deposit date: | 2016-10-03 | | Release date: | 2017-01-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal Structures of Human GlyRa3 Bound to a Novel Class of Potentiators with Efficacy in a Mouse Model of Neuropathic Pain
To Be Published
|
|
2MLH
 
 | | NMR Solution Structure of Opa60 from N. Gonorrhoeae in FC-12 Micelles | | Descriptor: | Opacity protein opA60 | | Authors: | Fox, D.A, Larsson, P, Lo, R.H, Kroncke, B.M, Kasson, P.M, Columbus, L. | | Deposit date: | 2014-02-27 | | Release date: | 2014-06-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the neisserial outer membrane protein opa60: loop flexibility essential to receptor recognition and bacterial engulfment.
J.Am.Chem.Soc., 136, 2014
|
|
2XUJ
 
 | | CRYSTAL STRUCTURE OF MACHE-Y337A-TZ2PA6 SYN COMPLEX (1 MTH) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,8-DIAMINO-6-PHENYL-5-[6-[1-[2-[(1,2,3,4-TETRAHYDRO-9-ACRIDINYL)AMINO]ETHYL]-1H-1,2,3-TRIAZOL-5-YL]HEXYL]-PHENANTHRIDINIUM, ACETYLCHOLINESTERASE, ... | | Authors: | Bourne, Y, Radic, Z, Taylor, P, Marchot, P. | | Deposit date: | 2010-10-19 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Conformational Remodeling of Femtomolar Inhibitor-Acetylcholinesterase Complexes in the Crystalline State
J.Am.Chem.Soc., 132, 2010
|
|
5KUJ
 
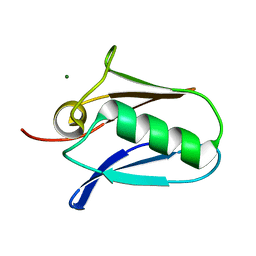 | | Human mitochondrial calcium uniporter (residues 72-189) crystal structure with magnesium. | | Descriptor: | Calcium uniporter protein, mitochondrial, MAGNESIUM ION | | Authors: | Mok, M.C.Y, Lee, S.K, Junop, M.S, Stathopulos, P.B. | | Deposit date: | 2016-07-13 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Insights into Mitochondrial Calcium Uniporter Regulation by Divalent Cations.
Cell Chem Biol, 23, 2016
|
|
5L6E
 
 | |
2LV0
 
 | |
3W5I
 
 | |
2TPR
 
 | |
2RU4
 
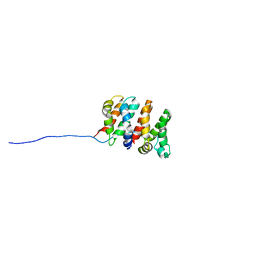 | | Designed Armadillo Repeat Protein Self-ASsembled Complex (YIIM2-MAII) | | Descriptor: | Armadillo Repeat Protein, C-terminal fragment, MAII, ... | | Authors: | Zerbe, O, Christen, M.T, Plueckthun, A, Watson, R.P. | | Deposit date: | 2013-11-22 | | Release date: | 2014-07-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Spontaneous self-assembly of engineered armadillo repeat protein fragments into a folded structure
Structure, 22, 2014
|
|
5WGG
 
 | | Structural Insights into Thioether Bond Formation in the Biosynthesis of Sactipeptides | | Descriptor: | CALCIUM ION, CteA, IRON/SULFUR CLUSTER, ... | | Authors: | Grove, T.L, Himes, P, Bowers, A, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2017-07-14 | | Release date: | 2017-07-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.036 Å) | | Cite: | Structural Insights into Thioether Bond Formation in the Biosynthesis of Sactipeptides.
J. Am. Chem. Soc., 139, 2017
|
|
1B5M
 
 | | RAT OUTER MITOCHONDRIAL MEMBRANE CYTOCHROME B5 | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Rivera, M, White, S.P, Zhang, X. | | Deposit date: | 1996-11-07 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 13C NMR spectroscopic and X-ray crystallographic study of the role played by mitochondrial cytochrome b5 heme propionates in the electrostatic binding to cytochrome c.
Biochemistry, 35, 1996
|
|
1AQ1
 
 | | HUMAN CYCLIN DEPENDENT KINASE 2 COMPLEXED WITH THE INHIBITOR STAUROSPORINE | | Descriptor: | CYCLIN-DEPENDENT PROTEIN KINASE 2, STAUROSPORINE | | Authors: | Endicott, J.A, Noble, M.E.M, Johnson, L.N, Lawrie, A, Tunnah, P, Brown, N.R. | | Deposit date: | 1997-08-05 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protein kinase inhibition by staurosporine revealed in details of the molecular interaction with CDK2.
Nat.Struct.Biol., 4, 1997
|
|
5LB3
 
 | | Crystal structure of human RECQL5 helicase in complex with ADP/Mg. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase Q5, MAGNESIUM ION, ... | | Authors: | Newman, J.A, Aitkenhead, H, Savitsky, P, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-15 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into the RecQ helicase mechanism revealed by the structure of the helicase domain of human RECQL5.
Nucleic Acids Res., 45, 2017
|
|
7QFP
 
 | | Cryo-EM structure of Botulinum neurotoxin serotype E | | Descriptor: | Botulinum neurotoxin | | Authors: | Kosenina, S, Martinez-Carranza, M, Davies, J.R, Masuyer, G, Stenmark, P. | | Deposit date: | 2021-12-06 | | Release date: | 2022-01-26 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural Analysis of Botulinum Neurotoxins Type B and E by Cryo-EM.
Toxins, 14, 2021
|
|
2XUK
 
 | | CRYSTAL STRUCTURE OF MACHE-Y337A-TZ2PA6 SYN COMPLEX (10 MTH) | | Descriptor: | 3,8-DIAMINO-6-PHENYL-5-[6-[1-[2-[(1,2,3,4-TETRAHYDRO-9-ACRIDINYL)AMINO]ETHYL]-1H-1,2,3-TRIAZOL-5-YL]HEXYL]-PHENANTHRIDINIUM, ACETYLCHOLINESTERASE | | Authors: | Bourne, Y, Radic, Z, Taylor, P, Marchot, P. | | Deposit date: | 2010-10-19 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Conformational Remodeling of Femtomolar Inhibitor-Acetylcholinesterase Complexes in the Crystalline State
J.Am.Chem.Soc., 132, 2010
|
|
1AOS
 
 | |
5T90
 
 | | Structural mechanisms for alpha-conotoxin selectivity at the human alpha3beta4 nicotinic acetylcholine receptor | | Descriptor: | Acetylcholine-binding protein, LsIA | | Authors: | Abraham, N, Healy, M, Ragnarsson, L, Brust, A, Alewood, P, Lewis, R. | | Deposit date: | 2016-09-08 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural mechanisms for alpha-conotoxin activity at the human alpha 3 beta 4 nicotinic acetylcholine receptor.
Sci Rep, 7, 2017
|
|
1BDY
 
 | | C2 DOMAIN FROM PROTEIN KINASE C DELTA | | Descriptor: | PROTEIN KINASE C | | Authors: | Pappa, H, Murray-Rust, J, Dekker, L.V, Parker, P.J, Mcdonald, N.Q. | | Deposit date: | 1998-05-11 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the C2 domain from protein kinase C-delta.
Structure, 6, 1998
|
|
1BIB
 
 | |
