8UOA
 
 | | Structure of the synaptic vesicle protein 2A Luminal domain in complex with a nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody, Synaptic vesicle glycoprotein 2A, ... | | Authors: | Mittal, A, Martin, M.F, Levin, E, Adams, C, Yang, M, Ledecq, M, Horanyi, P.S, Coleman, J.A. | | Deposit date: | 2023-10-19 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of synaptic vesicle protein 2A and 2B bound to anticonvulsants.
Nat.Struct.Mol.Biol., 2024
|
|
6MEG
 
 | | Crystal structure of human monoclonal antibody HEPC46 | | Descriptor: | antibody HEPC46 Heavy Chain, antibody HEPC46 Light Chain | | Authors: | Flyak, A.I, Bjorkman, P.J. | | Deposit date: | 2018-09-06 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | HCV Broadly Neutralizing Antibodies Use a CDRH3 Disulfide Motif to Recognize an E2 Glycoprotein Site that Can Be Targeted for Vaccine Design.
Cell Host Microbe, 24, 2018
|
|
8V4Q
 
 | | Myxococcus xanthus EncA 3xHis pore mutant with tetrahedral symmetry | | Descriptor: | Type 1 encapsulin shell protein EncA | | Authors: | Szyszka, T.N, Andreas, M.P, Lie, F, Miller, L.M, Adamson, L.S.R, Fatehi, F, Twarock, R, Draper, B.E, Jarrold, M.F, Giessen, T.W, Lau, Y.H. | | Deposit date: | 2023-11-29 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Point mutation in a virus-like capsid drives symmetry reduction to form tetrahedral cages.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8U3E
 
 | |
7XPE
 
 | | Cryo-EM structure of the T=4 lake sinai virus 2 virus-like capsid at pH 8.5 | | Descriptor: | Capsid protein alpha | | Authors: | Chen, N.C, Wang, C.H, Chen, C.J, Yoshimura, M, Guan, H.H, Chuankhayan, P, Lin, C.C. | | Deposit date: | 2022-05-04 | | Release date: | 2023-02-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structures of honeybee-infecting Lake Sinai virus reveal domain functions and capsid assembly with dynamic motions.
Nat Commun, 14, 2023
|
|
6F2Q
 
 | | Neutron crystal structure of perdeuterated galectin-3C in the ligand-free form | | Descriptor: | Galectin-3 | | Authors: | Manzoni, F, Blakeley, M.P, Oksanen, E, Logan, D.T. | | Deposit date: | 2017-11-27 | | Release date: | 2018-05-02 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (1.03 Å), X-RAY DIFFRACTION | | Cite: | Elucidation of Hydrogen Bonding Patterns in Ligand-Free, Lactose- and Glycerol-Bound Galectin-3C by Neutron Crystallography to Guide Drug Design.
J. Med. Chem., 61, 2018
|
|
7XGZ
 
 | | Cryo-EM structure of the T=4 lake sinai virus 2 virus-like capsid at pH 7.5 | | Descriptor: | Capsid protein alpha | | Authors: | Chen, N.C, Wang, C.H, Chen, C.J, Yoshimura, M, Guan, H.H, Chuankhayan, P, Lin, C.C. | | Deposit date: | 2022-04-07 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structures of honeybee-infecting Lake Sinai virus reveal domain functions and capsid assembly with dynamic motions
Nat Commun, 14, 2023
|
|
5EL9
 
 | | A. thaliana IGPD2 in complex with the triazole-phosphonate inhibitor, (S)-C348, to 1.1A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Imidazoleglycerol-phosphate dehydratase 2, ... | | Authors: | Bisson, C, Britton, K.L, Sedelnikova, S.E, Rodgers, H.F, Eadsforth, T.C, Viner, R.C, Hawkes, T.R, Baker, P.J, Rice, D.W. | | Deposit date: | 2015-11-04 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Mirror-Image Packing Provides a Molecular Basis for the Nanomolar Equipotency of Enantiomers of an Experimental Herbicide.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
7XPA
 
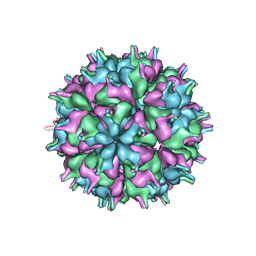 | | Cryo-EM structure of the T=3 lake sinai virus 2 virus-like capsid at pH 7.5 | | Descriptor: | Capsid protein alpha | | Authors: | Chen, N.C, Wang, C.H, Chen, C.J, Yoshimura, M, Guan, H.H, Chuankhayan, P, Lin, C.C. | | Deposit date: | 2022-05-04 | | Release date: | 2023-02-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structures of honeybee-infecting Lake Sinai virus reveal domain functions and capsid assembly with dynamic motions.
Nat Commun, 14, 2023
|
|
7XPB
 
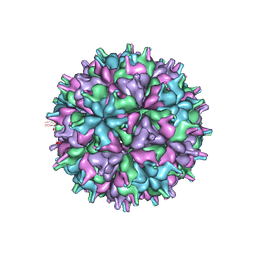 | | Cryo-EM structure of the T=4 lake sinai virus 2 virus-like capsid at pH 6.5 | | Descriptor: | Capsid protein alpha | | Authors: | Chen, N.C, Wang, C.H, Chen, C.J, Yoshimura, M, Guan, H.H, Chuankhayan, P, Lin, C.C. | | Deposit date: | 2022-05-04 | | Release date: | 2023-02-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Structures of honeybee-infecting Lake Sinai virus reveal domain functions and capsid assembly with dynamic motions.
Nat Commun, 14, 2023
|
|
5EFV
 
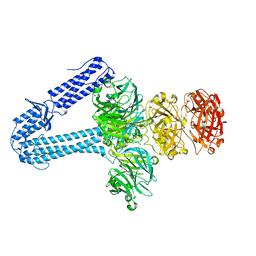 | | The host-recognition device of Staphylococcus aureus phage Phi11 | | Descriptor: | FE (III) ION, Phi ETA orf 56-like protein | | Authors: | Koc, C, Kuhner, P, Xia, G, Spinelli, S, Roussel, A, Cambillau, C, Stehle, T. | | Deposit date: | 2015-10-26 | | Release date: | 2016-05-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the host-recognition device of Staphylococcus aureus phage 11.
Sci Rep, 6, 2016
|
|
5NJM
 
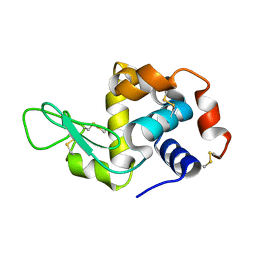 | | Lysozyme room-temperature structure determined by serial millisecond crystallography | | Descriptor: | Lysozyme C | | Authors: | Weinert, T, Vera, L, Marsh, M, James, D, Gashi, D, Nogly, P, Jaeger, K, Standfuss, J. | | Deposit date: | 2017-03-29 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons.
Nat Commun, 8, 2017
|
|
8BXP
 
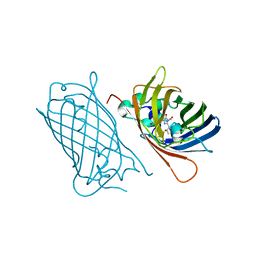 | |
8U3D
 
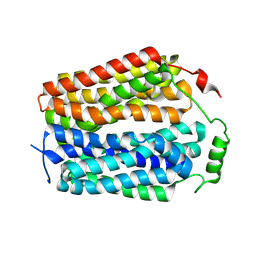 | |
6HV2
 
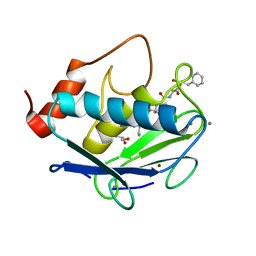 | | MMP-13 in complex with the peptide IMISF | | Descriptor: | CALCIUM ION, Collagenase 3, GLYCEROL, ... | | Authors: | Mittl, P, Riedl, R, Hohl, D. | | Deposit date: | 2018-10-10 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.709 Å) | | Cite: | Drug Design Inspired by Nature: Crystallographic Detection of an Auto-Tailored Protease Inhibitor Template.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
4QGO
 
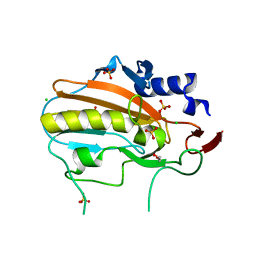 | | Crystal structure of NucA from Streptococcus agalactiae with no metal bound | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA-entry nuclease (Competence-specific nuclease), ... | | Authors: | Pedersen, L.C, Moon, A.F, Gaudu, P. | | Deposit date: | 2014-05-23 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural characterization of the virulence factor nuclease A from Streptococcus agalactiae.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7XF3
 
 | | The structure of HLA-B*1501/BM58-66AF9 | | Descriptor: | 9-mer peptide from Matrix protein 1, Beta-2-microglobulin, MHC class I antigen | | Authors: | Zhao, Y.Z, Xiao, W.L, Wu, Y.N, Fan, W.F, Yue, C, Zhang, Q.X, Zhang, D.N, Yuan, X.J, Yao, S.J, Liu, S, Li, M, Wang, P.Y, Zhang, H.J, Zhang, J, Zhao, M, Zheng, X.Q, Liu, W.J, Gao, G.F, Liu, W.L. | | Deposit date: | 2022-03-31 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Parallel T Cell Immunogenic Regions in Influenza B and A Viruses with Distinct Nuclear Export Signal Functions: The Balance between Viral Life Cycle and Immune Escape.
J Immunol., 210, 2023
|
|
6VL1
 
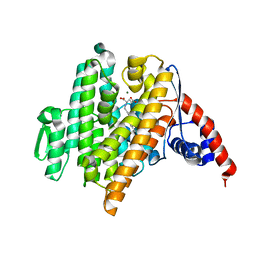 | | Crystal Structure of the N-prenyltransferase DabA in Complex with NGG and Mg2+ | | Descriptor: | DabA, MAGNESIUM ION, N-[(2E)-3,7-dimethylocta-2,6-dien-1-yl]-L-glutamic acid | | Authors: | Chekan, J.R, Noel, J.P, Moore, B.S. | | Deposit date: | 2020-01-22 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Algal neurotoxin biosynthesis repurposes the terpene cyclase structural fold into anN-prenyltransferase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5EP5
 
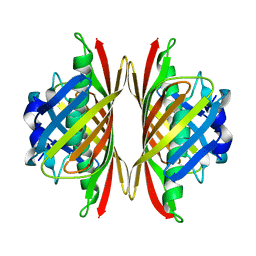 | |
3R2H
 
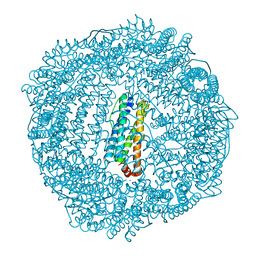 | | 1.7 A resolution structure of As-Isolated FtnA from Pseudomonas aeruginosa (pH 10.5) | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Bacterioferritin, SODIUM ION | | Authors: | Lovell, S.W, Battaile, K.P, Yao, H, Jepkorir, G, Nama, P.V, Weeratunga, S, Rivera, M. | | Deposit date: | 2011-03-14 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Two distinct ferritin-like molecules in Pseudomonas aeruginosa: the product of the bfrA gene is a bacterial ferritin (FtnA) and not a bacterioferritin (Bfr).
Biochemistry, 50, 2011
|
|
8U3G
 
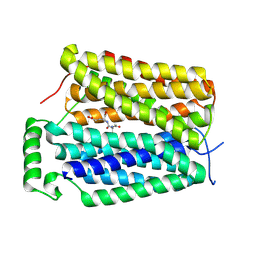 | | Structure of NAAG-bound Sialin | | Descriptor: | ACETYL GROUP, ASPARTIC ACID, GLUTAMIC ACID, ... | | Authors: | Schmiege, P, Li, X. | | Deposit date: | 2023-09-07 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Structure of NAAG-bound Sialin
To Be Published
|
|
3FRL
 
 | | The 2.25 A crystal structure of LipL32, the major surface antigen of Leptospira interrogans serovar Copenhageni | | Descriptor: | 2,2',2''-NITRILOTRIETHANOL, CHLORIDE ION, LipL32, ... | | Authors: | Farah, C.S, Guzzo, C.R, Hauk, P, Ho, P.L. | | Deposit date: | 2009-01-08 | | Release date: | 2009-06-16 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure and calcium-binding activity of LipL32, the major surface antigen of pathogenic Leptospira sp.
J.Mol.Biol., 390, 2009
|
|
5FYY
 
 | | Crystal structure of the catalytic domain of human JARID1B in complex with Maybridge fragment 3-pyridin-3-ylaniline (N05798a) (ligand modelled based on PANDDA event map) | | Descriptor: | 1,2-ETHANEDIOL, 3-(pyridin-3-yl)aniline, CHLORIDE ION, ... | | Authors: | Nowak, R, Krojer, T, Johansson, C, Gileadi, C, Kupinska, K, Strain-Damerell, C, Szykowska, A, Pearce, N, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, von Delft, F, Brennan, P.E, Oppermann, U. | | Deposit date: | 2016-03-10 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Jarid1B in Complex with N05798A
To be Published
|
|
4GW8
 
 | | Human proto-oncogene serine threonine kinase (PIM1) in complex with a consensus peptide and Leucettine L41 | | Descriptor: | 1,2-ETHANEDIOL, 5-(1,3-benzodioxol-5-ylmethyl)-2-(phenylamino)-4H-imidazol-4-one, Consensus peptide (Pimtide), ... | | Authors: | Filippakopoulos, P, Bullock, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Meijer, L, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-09-01 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selectivity, cocrystal structures, and neuroprotective properties of leucettines, a family of protein kinase inhibitors derived from the marine sponge alkaloid leucettamine B.
J.Med.Chem., 55, 2012
|
|
6MCX
 
 | | INFLUENZA VIRUS NEURAMINIDASE SUBTYPE N9 (TERN) RECOMBINANT HEAD DOMAIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Streltsov, V.A, Schmidt, P, McKimm-Breschkin, J. | | Deposit date: | 2018-09-02 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of an Influenza A virus N9 neuraminidase with a tetrabrachion-domain stalk.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
