5DYY
 
 | | Crystal structure of human butyrylcholinesterase in complex with N-((1-benzylpiperidin-3-yl)methyl)naphthalene-2-sulfonamide | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Coquelle, N, Brus, B, Colletier, J.P. | | Deposit date: | 2015-09-25 | | Release date: | 2017-01-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Development of an in-vivo active reversible butyrylcholinesterase inhibitor.
Sci Rep, 6, 2016
|
|
3U4A
 
 | | From soil to structure: a novel dimeric family 3-beta-glucosidase isolated from compost using metagenomic analysis | | Descriptor: | CALCIUM ION, JMB19063, beta-D-glucopyranose, ... | | Authors: | McAndrew, R.P, Park, J.I, Reindl, W, Friedland, G.D, D'haeseleer, P, Northen, T, Sale, K.L, Simmons, B.A, Adams, P.D. | | Deposit date: | 2011-10-07 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | From soil to structure: a novel dimeric family 3-beta--glucosidase isolated from compost using metagenomic analysis
To be Published
|
|
6GRP
 
 | | Crystal Structure Of Human Transthyretin in complex with 3,5,6-trichloro-2-pyridinol (TC2P) | | Descriptor: | 3,5,6-trichloro-2-pyridinol, SODIUM ION, Transthyretin | | Authors: | Grundstrom, C, Zhang, J, Olofsson, A, Andersson, P.L, Sauer-Eriksson, A.E. | | Deposit date: | 2018-06-12 | | Release date: | 2018-07-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Interspecies Variation between Fish and Human Transthyretins in Their Binding of Thyroid-Disrupting Chemicals.
Environ. Sci. Technol., 52, 2018
|
|
2XFE
 
 | | vCBM60 in complex with galactobiose | | Descriptor: | CALCIUM ION, CARBOHYDRATE BINDING MODULE, beta-D-galactopyranose-(1-4)-beta-D-galactopyranose | | Authors: | Montanier, C, Flint, J.E, Bolam, D.N, Xie, H, Liu, Z, Rogowski, A, Weiner, D.P, Nurizzo, D, Roberts, S.M, Turkenburg, J.P, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2010-05-21 | | Release date: | 2010-06-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Circular Permutation Provides an Evolutionary Link between Two Families of Calcium-Dependent Carbohydrate Binding Modules.
J.Biol.Chem., 285, 2010
|
|
5DOW
 
 | |
6GEZ
 
 | | THE STRUCTURE OF TWITCH-2B N532F | | Descriptor: | CALCIUM ION, FORMIC ACID, Green fluorescent protein,Optimized Ratiometric Calcium Sensor,Green fluorescent protein,Green fluorescent protein | | Authors: | Trigo Mourino, P, Paulat, M, Thestrup, T, Griesbeck, O, Griesinger, C, Becker, S. | | Deposit date: | 2018-04-27 | | Release date: | 2019-08-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Dynamic tuning of FRET in a green fluorescent protein biosensor.
Sci Adv, 5, 2019
|
|
2XFK
 
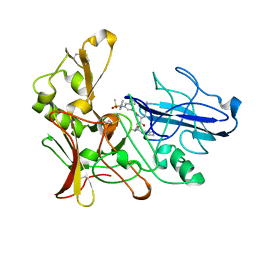 | | Human BACE-1 in complex with N-((1S,2R)-3-(((1S)-2-(cyclohexylamino)- 1-methyl-2-oxoethyl)amino)-2-hydroxy-1-(phenylmethyl)propyl)-3-(ethylamino)-5-((methylsulfonyl)(phenyl)amino)benzamide | | Descriptor: | BETA-SECRETASE 1, N-((1S,2R)-3-(((1S)-2-(CYCLOHEXYLAMINO)-1--METHYL-2-OXOETHYL)AMINO)-2-HYDROXY-1-(PHENYLMETHYL)PROPYL)-3-(ETHYLAMINO)-5-((METHYLSULFONYL)(PHENYL)AMINO)BENZAMIDE | | Authors: | Clarke, B, Cutler, L, Demont, E, Dingwall, C, Dunsdon, R, Hawkins, J, Howes, C, Hussain, I, Maile, G, Matico, R, Mosley, J, Naylor, A, O'Brien, A, Redshaw, S, Rowland, P, Soleil, V, Smith, K.J, Sweitzer, S, Theobald, P, Vesey, D, Walter, D.S, Wayne, G. | | Deposit date: | 2010-05-24 | | Release date: | 2010-07-07 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bace-1 Inhibitors Using Novel Edge-to-Face Interaction with Arg-296
Bioorg.Med.Chem.Lett., 20, 2010
|
|
4GWL
 
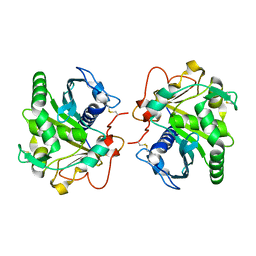 | | Structure of three phase partition treated lipase from Thermomyces lanuginosa at 2.55A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lipase | | Authors: | Shukla, P.K, Sinha, M, Mukherjee, J, Gupta, M.N, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-09-03 | | Release date: | 2012-09-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of three phase partition treated lipase from Thermomyces lanuginosa at 2.55A resolution
To be Published
|
|
5JP4
 
 | |
8EFS
 
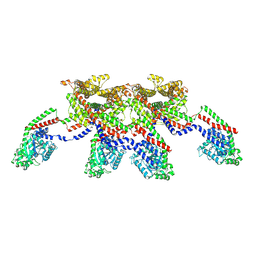 | | CryoEM of the soluble OPA1 tetramer from the apo helical assembly on a lipid membrane | | Descriptor: | Dynamin-like 120 kDa protein, form S1 | | Authors: | Nyenhuis, S.B, Wu, X, Stanton, A.E, Strub, M.P, Yim, Y.I, Canagarajah, B, Hinshaw, J.E. | | Deposit date: | 2022-09-09 | | Release date: | 2023-06-28 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (9.68 Å) | | Cite: | OPA1 helical structures give perspective to mitochondrial dysfunction.
Nature, 620, 2023
|
|
7UNA
 
 | | SfSTING with cGAMP (masked) | | Descriptor: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, CD-NTase-associated protein 12 | | Authors: | Morehouse, B.R, Yip, M.C.J, Keszei, A.F.A, McNamara-Bordewick, N.K, Shao, S, Kranzusch, P.J. | | Deposit date: | 2022-04-09 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of an active bacterial TIR-STING filament complex.
Nature, 608, 2022
|
|
1A35
 
 | | HUMAN TOPOISOMERASE I/DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*AP*GP*AP*CP*TP*TP*AP*GP*AP*AP*AP*AP*AP*(BRU)P*(BRU)P*TP*TP*T)-3'), DNA (5'-D(*AP*AP*AP*AP*AP*TP*+UP*+UP*+UP*+UP*CP*+UP*AP*AP*GP*TP*CP*TP*TP*TP*+ UP*T)-3'), PROTEIN (DNA TOPOISOMERASE I) | | Authors: | Redinbo, M.R, Stewart, L, Kuhn, P, Champoux, J.J, Hol, W.G. | | Deposit date: | 1998-01-29 | | Release date: | 1998-08-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA.
Science, 279, 1998
|
|
8EEW
 
 | | CryoEM of the soluble OPA1 dimer from the GDP-AlFx bound helical assembly on a lipid membrane | | Descriptor: | Dynamin-like 120 kDa protein, form S1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Nyenhuis, S.B, Wu, X, Stanton, A.E, Strub, M.P, Yim, Y.I, Canagarajah, B, Hinshaw, J.E. | | Deposit date: | 2022-09-07 | | Release date: | 2023-06-28 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (5.48 Å) | | Cite: | OPA1 helical structures give perspective to mitochondrial dysfunction.
Nature, 620, 2023
|
|
7F03
 
 | | Cytochrome c-type biogenesis protein CcmABCD from E. coli in complex with ANP | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, ... | | Authors: | Li, J, Zheng, W, Gu, M, Zhang, K, Zhu, J.P. | | Deposit date: | 2021-06-03 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
8EF7
 
 | | CryoEM of the soluble OPA1 dimer from the apo helical assembly on a lipid membrane | | Descriptor: | Dynamin-like 120 kDa protein, form S1 | | Authors: | Nyenhuis, S.B, Wu, X, Stanton, A.E, Strub, M.P, Yim, Y.I, Canagarajah, B, Hinshaw, J.E. | | Deposit date: | 2022-09-08 | | Release date: | 2023-06-28 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (9.68 Å) | | Cite: | OPA1 helical structures give perspective to mitochondrial dysfunction.
Nature, 620, 2023
|
|
7F04
 
 | | Cytochrome c-type biogenesis protein CcmABCD from E. coli in complex with Heme and ATP. | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, ADENOSINE-5'-TRIPHOSPHATE, Cytochrome c biogenesis ATP-binding export protein CcmA, ... | | Authors: | Li, J, Zheng, W, Gu, M, Zhang, K, Zhu, J.P. | | Deposit date: | 2021-06-03 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
2HA7
 
 | | Crystal structure of mutant S203A of mouse acetylcholinesterase complexed with butyrylthiocholine | | Descriptor: | 2-(BUTYRYLSULFANYL)-N,N,N-TRIMETHYLETHANAMINIUM, 2-(TRIMETHYLAMMONIUM)ETHYL THIOL, Acetylcholinesterase, ... | | Authors: | Bourne, Y, Radic, Z, Sulzenbacher, G, Kim, E, Taylor, P, Marchot, P. | | Deposit date: | 2006-06-12 | | Release date: | 2006-07-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Substrate and product trafficking through the active center gorge of acetylcholinesterase analyzed by crystallography and equilibrium binding
J.Biol.Chem., 281, 2006
|
|
3PAL
 
 | | IONIC INTERACTIONS WITH PARVALBUMINS. CRYSTAL STRUCTURE DETERMINATION OF PIKE 4.10 PARVALBUMIN IN FOUR DIFFERENT IONIC ENVIRONMENTS | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PARVALBUMIN | | Authors: | Declercq, J.P, Tinant, B, Parello, J, Rambaud, J. | | Deposit date: | 1990-11-08 | | Release date: | 1992-01-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Ionic interactions with parvalbumins. Crystal structure determination of pike 4.10 parvalbumin in four different ionic environments.
J.Mol.Biol., 220, 1991
|
|
4ENR
 
 | | Structure of E530I variant E. coli KatE | | Descriptor: | Catalase HPII, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Loewen, P.C, Jha, V. | | Deposit date: | 2012-04-13 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Influence of main channel structure on H(2)O(2) access to the heme cavity of catalase KatE of Escherichia coli.
Arch.Biochem.Biophys., 526, 2012
|
|
4AOB
 
 | | SAM-I riboswitch containing the T. solenopsae Kt-23 in complex with S- adenosyl methionine | | Descriptor: | BARIUM ION, POTASSIUM ION, S-ADENOSYLMETHIONINE, ... | | Authors: | Schroeder, K.T, Daldrop, P, McPhee, S.A, Lilley, D.M.J. | | Deposit date: | 2012-03-25 | | Release date: | 2012-05-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure and Folding of a Rare, Natural Kink Turn in RNA with an Aa Pair at the 2B2N Position.
RNA, 18, 2012
|
|
6GNO
 
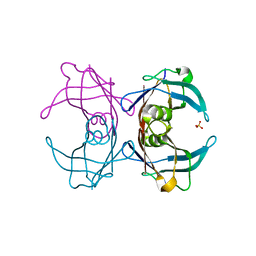 | | Crystal Structure Of Sea Bream Transthyretin in complex with Tetrabromobisphenol A (TBBPA) | | Descriptor: | 4,4'-propane-2,2-diylbis(2,6-dibromophenol), SULFATE ION, Transthyretin | | Authors: | Grundstrom, C, Zhang, J, Olofsson, A, Andersson, P.L, Sauer-Eriksson, A.E. | | Deposit date: | 2018-05-31 | | Release date: | 2018-07-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Interspecies Variation between Fish and Human Transthyretins in Their Binding of Thyroid-Disrupting Chemicals.
Environ. Sci. Technol., 52, 2018
|
|
3HD4
 
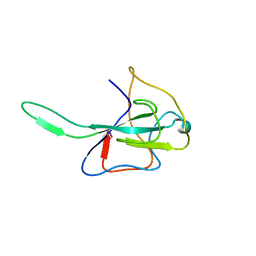 | | MHV Nucleocapsid Protein NTD | | Descriptor: | Nucleoprotein | | Authors: | Giedroc, D.P, Keane, S.C, Dann III, C.E. | | Deposit date: | 2009-05-06 | | Release date: | 2009-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.747 Å) | | Cite: | Coronavirus N Protein N-Terminal Domain (NTD) Specifically Binds the Transcriptional Regulatory Sequence (TRS) and Melts TRS-cTRS RNA Duplexes.
J.Mol.Biol., 394, 2009
|
|
2CLX
 
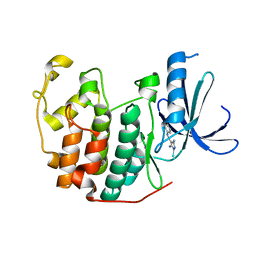 | | 4-Arylazo-3,5-diamino-1H-pyrazole CDK Inhibitors: SAR Study, Crystal Structure in Complex with CDK2, Selectivity, and Cellular Effects | | Descriptor: | 4-[(E)-(3,5-DIAMINO-1H-PYRAZOL-4-YL)DIAZENYL]PHENOL, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Krystof, V, Cankar, P, Frysova, I, Slouka, J, Kontopidis, G, Dzubak, P, Hajduch, M, Deazevedo, W.F, Paprskarova, M, Orsag, M, Rolcik, J, Latr, A, Fischer, P.M, Strnad, M. | | Deposit date: | 2006-05-02 | | Release date: | 2006-11-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 4-Arylazo-3,5-Diamino-1H-Pyrazole Cdk Inhibitors: Sar Study, Crystal Structure in Complex with Cdk2, Selectivity, and Cellular Effects
J.Med.Chem., 49, 2006
|
|
5EAR
 
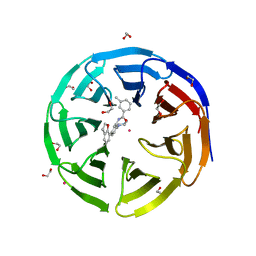 | | Crystal structure of human WDR5 in complex with compound 9d | | Descriptor: | 1,2-ETHANEDIOL, N-[5-(2,3-dihydro-1-benzofuran-7-yl)-2-(4-methylpiperazin-1-yl)phenyl]-3-methylbenzamide, SULFATE ION, ... | | Authors: | DONG, A, DOMBROVSKI, L, SMIL, D, GETLIK, M, BOLSHAN, Y, WALKER, J.R, SENISTERRA, G, PODA, G, AL-AWAR, R, SCHAPIRA, M, VEDADI, M, Bountra, C, Edwards, A.M, Arrowsmith, C.H, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-10-16 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Optimization of a Small Molecule Antagonist of the Interaction Between WD Repeat-Containing Protein 5 (WDR5) and Mixed-Lineage Leukemia 1 (MLL1).
J. Med. Chem., 59, 2016
|
|
1Y6T
 
 | | HIV-1 Dis(Mal) Duplex Co Hexamine-Soaked | | Descriptor: | 5'-R(*CP*UP*UP*GP*CP*UP*GP*AP*GP*GP*UP*GP*CP*AP*CP*AP*CP*AP*GP*CP*AP*AP*G)-3', COBALT (III) ION, SODIUM ION | | Authors: | Ennifar, E, Walter, P, Dumas, P. | | Deposit date: | 2004-12-07 | | Release date: | 2004-12-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A crystallographic study of the binding of 13 metal ions to two related RNA duplexes
Nucleic Acids Res., 31, 2003
|
|
