7YF0
 
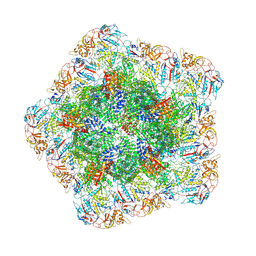 | | In situ structure of polymerase complex of mammalian reovirus in the core | | Descriptor: | Lambda-2 protein, Mu-2 protein, RNA helicase, ... | | Authors: | Bao, K.Y, Zhang, X.L, Li, D.Y, Zhu, P. | | Deposit date: | 2022-07-07 | | Release date: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | In situ structures of polymerase complex of mammalian reovirus illuminate RdRp activation and transcription regulation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6DWG
 
 | |
7YEV
 
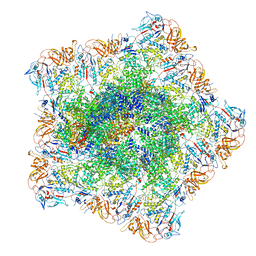 | | In situ structure of polymerase complex of mammalian reovirus in the pre-elongation state | | Descriptor: | Lambda-2 protein, Mu-2 protein, RNA helicase, ... | | Authors: | Bao, K.Y, Zhang, X.L, Li, D.Y, Zhu, P. | | Deposit date: | 2022-07-06 | | Release date: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | In situ structures of polymerase complex of mammalian reovirus illuminate RdRp activation and transcription regulation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5E89
 
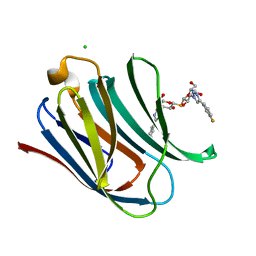 | | Crystal structure of Human galectin-3 CRD in complex with 3-fluophenyl-1,2,3-triazolyl thiodigalactoside inhibitor | | Descriptor: | 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-beta-D-galactopyranosyl 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-1-thio-beta-D-galactopyranoside, CHLORIDE ION, Galectin-3 | | Authors: | Collins, P.M, Blanchard, H. | | Deposit date: | 2015-10-13 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Galectin-3-Binding Glycomimetics that Strongly Reduce Bleomycin-Induced Lung Fibrosis and Modulate Intracellular Glycan Recognition.
Chembiochem, 17, 2016
|
|
5E8A
 
 | | Crystal structure of Human galectin-3 CRD in complex with 4-fluophenyl-1,2,3-triazolyl thiodigalactoside inhibitor | | Descriptor: | 3-deoxy-3-[4-(4-fluorophenyl)-1H-1,2,3-triazol-1-yl]-beta-D-galactopyranosyl 3-deoxy-3-[4-(4-fluorophenyl)-1H-1,2,3-triazol-1-yl]-1-thio-beta-D-galactopyranoside, CHLORIDE ION, Galectin-3 | | Authors: | Collins, P.M, Blanchard, H. | | Deposit date: | 2015-10-13 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Galectin-3-Binding Glycomimetics that Strongly Reduce Bleomycin-Induced Lung Fibrosis and Modulate Intracellular Glycan Recognition.
Chembiochem, 17, 2016
|
|
6DQH
 
 | | Cronobacter sakazakii (Enterobacter sakazakii) Metallo-beta-lactamse HARLDQ motif | | Descriptor: | Beta-lactamase, PHOSPHATE ION, ZINC ION | | Authors: | Monteiro Pedroso, M, Waite, D, Natasa, M, McGeary, R, Guddat, L, Hugenholtz, P, Schenk, G. | | Deposit date: | 2018-06-11 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.104 Å) | | Cite: | Broad spectrum antibiotic-degrading metallo-beta-lactamases are phylogenetically diverse
Protein Cell, 2020
|
|
2GF0
 
 | | The crystal structure of the human DiRas1 GTPase in the inactive GDP bound state | | Descriptor: | GTP-binding protein Di-Ras1, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Turnbull, A.P, Papagrigoriou, E, Yang, X, Schoch, G, Elkins, J, Gileadi, O, Salah, E, Bray, J, Wen-Hwa, L, Fedorov, O, Niesen, F.E, von Delft, F, Weigelt, J, Edwards, A, Arrowsmith, C, Sundstrom, M, Doyle, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-03-21 | | Release date: | 2006-04-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of the human DiRas1 GTPase in the inactive GDP bound state
To be Published
|
|
8E4A
 
 | | Pseudomonas LpxC in complex with LPC-233 | | Descriptor: | 4-(4-cyclopropylbuta-1,3-diyn-1-yl)-N-[(2S,3S)-4,4-difluoro-3-hydroxy-1-(hydroxyamino)-3-methyl-1-oxobutan-2-yl]benzamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Najeeb, J, Zhou, P. | | Deposit date: | 2022-08-17 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.034 Å) | | Cite: | Preclinical safety and efficacy characterization of an LpxC inhibitor against Gram-negative pathogens.
Sci Transl Med, 15, 2023
|
|
5EAP
 
 | | Crystal structure of human WDR5 in complex with compound 9e | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | DONG, A, DOMBROVSKI, L, SMIL, D, GETLIK, M, BOLSHAN, Y, WALKER, J.R, SENISTERRA, G, PODA, G, AL-AWAR, R, SCHAPIRA, M, VEDADI, M, Bountra, C, Edwards, A.M, Arrowsmith, C.H, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-10-16 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure-Based Optimization of a Small Molecule Antagonist of the Interaction Between WD Repeat-Containing Protein 5 (WDR5) and Mixed-Lineage Leukemia 1 (MLL1).
J. Med. Chem., 59, 2016
|
|
2G21
 
 | | Ketopiperazine-Based Renin Inhibitors: Optimization of the "C" Ring | | Descriptor: | 7-(2,4-DIAMINO-6-ETHYLPYRIMIDIN-5-YL)-1-(3-METHOXYPROPYL)QUINOLINIUM, Renin | | Authors: | Holsworth, D.D, Jalaiea, M, Zhanga, E, Mcconnella, P. | | Deposit date: | 2006-02-15 | | Release date: | 2006-06-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ketopiperazine-Based Renin Inhibitors: Optimization of the "C" Ring
BIOORG.MED.CHEM.LETT., 16, 2006
|
|
5DTF
 
 | |
2FZP
 
 | | Crystal structure of the USP8 interaction domain of human NRDP1 | | Descriptor: | ring finger protein 41 isoform 1 | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Newman, E.M, Butler-Cole, C, Finerty Jr, P.J, Weigelt, J, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-02-10 | | Release date: | 2006-03-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Amino-terminal Dimerization, NRDP1-Rhodanese Interaction, and Inhibited Catalytic Domain Conformation of the Ubiquitin-specific Protease 8 (USP8).
J.Biol.Chem., 281, 2006
|
|
2GGE
 
 | | Crystal Structure of Mandelate Racemase/Muconate Lactonizing Enzyme from Bacillus Subtilis complexed with MG++ at 1.8 A | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, yitF | | Authors: | Malashkevich, V.N, Sauder, J.M, Schwinn, K.D, Emtage, S, Thompson, D.A, Rutter, M.E, Dickey, M, Groshong, C, Bain, K.T, Adams, J.M, Reyes, C, Rooney, I, Powell, A, Boice, A, Gheyi, T, Ozyurt, S, Atwell, S, Wasserman, S.R, Burley, S.K, Sali, A, Babbitt, P, Pieper, U, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-03-23 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal Structure of Mandelate Racemase/Muconate Lactonizing Enzyme from Bacillus Subtilis complexed with MG++ at 1.8 A
To be Published
|
|
2GIN
 
 | |
8EYZ
 
 | | Engineered glutamine binding protein bound to GLN and a cobaloxime ligand | | Descriptor: | AZIDOBIS (DIMETHYLGLYOXIMATO) PYRIDINECOBALT, Amino acid ABC transporter substrate-binding protein, GLUTAMINE, ... | | Authors: | Bridwell-Rabb, J, Boggs, D.G, Olshansky, L, Fatima, S, Thompson, P. | | Deposit date: | 2022-10-29 | | Release date: | 2022-11-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Engineering a Conformationally Switchable Artificial Metalloprotein.
J.Am.Chem.Soc., 144, 2022
|
|
4V61
 
 | | Homology model for the Spinach chloroplast 30S subunit fitted to 9.4A cryo-EM map of the 70S chlororibosome. | | Descriptor: | 16S rRNA, 23S rRNA, 4.8S rRNA, ... | | Authors: | Sharma, M.R, Wilson, D.N, Datta, P.P, Barat, C, Schluenzen, F, Fucini, P, Agrawal, R.K. | | Deposit date: | 2007-11-09 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Cryo-EM study of the spinach chloroplast ribosome reveals the structural and functional roles of plastid-specific ribosomal proteins
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4V6T
 
 | | Structure of the bacterial ribosome complexed by tmRNA-SmpB and EF-G during translocation and MLD-loading | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Ramrath, D.J.F, Yamamoto, H, Rother, K, Wittek, D, Pech, M, Mielke, T, Loerke, J, Scheerer, P, Ivanov, P, Teraoka, Y, Shpanchenko, O, Nierhaus, K.H, Spahn, C.M.T. | | Deposit date: | 2012-01-27 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (8.3 Å) | | Cite: | The complex of tmRNA-SmpB and EF-G on translocating ribosomes.
Nature, 485, 2012
|
|
3H5L
 
 | | Crystal structure of a putative branched-chain amino acid ABC transporter from Silicibacter pomeroyi | | Descriptor: | putative Branched-chain amino acid ABC transporter | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Iizuka, M, Sampathkumar, P, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-22 | | Release date: | 2009-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a putative branched-chain amino acid ABC transporter from Silicibacter pomeroyi
To be Published
|
|
5VH3
 
 | | Crystal structure of Fab fragment of the anti-TNFa antibody infliximab in a C-centered orthorhombic crystal form | | Descriptor: | 1,2-ETHANEDIOL, Infliximab Fab Heavy Chain, Infliximab Fab Light Chain | | Authors: | Mayclin, S.J, Edwards, T.E, Lerch, T.F, Conlan, H, Sharpe, P. | | Deposit date: | 2017-04-12 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Infliximab crystal structures reveal insights into self-association.
MAbs, 9, 2017
|
|
1MK0
 
 | | catalytic domain of intron endonuclease I-TevI, E75A mutant | | Descriptor: | BETA-MERCAPTOETHANOL, CITRIC ACID, Intron-associated endonuclease 1 | | Authors: | Van Roey, P, Meehan, L, Kowalski, J.C, Belfort, M, Derbyshire, V. | | Deposit date: | 2002-08-28 | | Release date: | 2002-10-30 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Catalytic domain structure and hypothesis for function of GIY-YIG intron endonuclease I-TevI.
Nat.Struct.Biol., 9, 2002
|
|
4F3D
 
 | |
9EQK
 
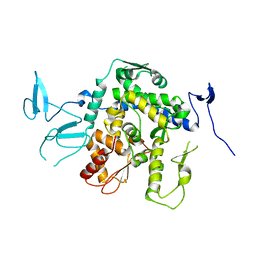 | |
1MG1
 
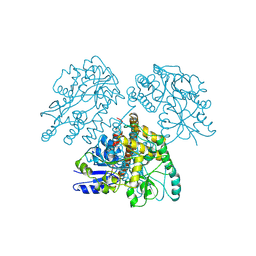 | | HTLV-1 GP21 ECTODOMAIN/MALTOSE-BINDING PROTEIN CHIMERA | | Descriptor: | CHLORIDE ION, PROTEIN (HTLV-1 GP21 ECTODOMAIN/MALTOSE-BINDING PROTEIN CHIMERA), alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Kobe, B, Center, R.J, Kemp, B.E, Poumbourios, P. | | Deposit date: | 1999-03-01 | | Release date: | 1999-04-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human T cell leukemia virus type 1 gp21 ectodomain crystallized as a maltose-binding protein chimera reveals structural evolution of retroviral transmembrane proteins.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
6JT7
 
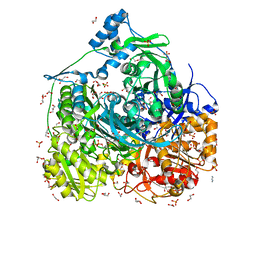 | | Crystal structure of 452-453_deletion mutant of FGAM Synthetase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Sharma, N, Ahalawat, N, Sandhu, P, Mondal, J, Anand, R. | | Deposit date: | 2019-04-10 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Role of allosteric switches and adaptor domains in long-distance cross-talk and transient tunnel formation.
Sci Adv, 6, 2020
|
|
3H33
 
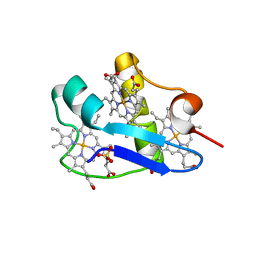 | | PpcC, A cytochrome c7 from Geobacter sulfurreducens | | Descriptor: | Cytochrome c7, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Pokkuluri, P.R, Schiffer, M. | | Deposit date: | 2009-04-15 | | Release date: | 2009-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural characterization of a family of cytochromes c(7) involved in Fe(III) respiration by Geobacter sulfurreducens.
Biochim.Biophys.Acta, 1797, 2010
|
|
