6MPC
 
 | |
4NOX
 
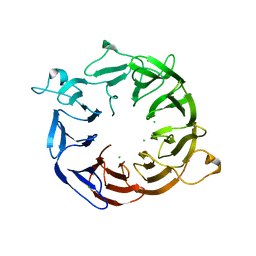 | | Structure of the nine-bladed beta-propeller of eIF3b | | Descriptor: | CHLORIDE ION, Eukaryotic translation initiation factor 3 subunit B | | Authors: | Liu, Y, Neumann, P, Kuhle, B, Monecke, T, Ficner, R. | | Deposit date: | 2013-11-20 | | Release date: | 2014-09-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.722 Å) | | Cite: | Translation initiation factor eIF3b contains a nine-bladed beta-propeller and interacts with the 40S ribosomal subunit
Structure, 22, 2014
|
|
6MPA
 
 | |
6MQ0
 
 | |
3LB4
 
 | | Two-site competitive inhibition in dehaloperoxidase-hemoglobin | | Descriptor: | 4-FLUOROPHENOL, Dehaloperoxidase A, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | de Serrano, V.S, Franzen, S, Thompson, M.K, Davis, M.F, Nicoletti, F.P, Howes, B.D, Smulevich, G. | | Deposit date: | 2010-01-07 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Internal binding of halogenated phenols in dehaloperoxidase-hemoglobin inhibits peroxidase function.
Biophys.J., 99, 2010
|
|
6MR6
 
 | |
4TJY
 
 | | Crystal Structure of human Tankyrase 2 in complex with ABT-888. | | Descriptor: | 2-[(2S)-2-methylpyrrolidin-2-yl]-1H-benzimidazole-7-carboxamide, Tankyrase-2, ZINC ION | | Authors: | Qiu, W, Lam, R, Romanov, V, Gordon, R, Gebremeskel, S, Vodsedalek, J, Thompson, C, Beletskaya, I, Battaile, K.P, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-05-25 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into the binding of PARP inhibitors to the catalytic domain of human tankyrase-2.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4TKP
 
 | | Complex of Ubc13 with the RING domain of the TRIM5alpha retroviral restriction factor | | Descriptor: | SULFATE ION, Tripartite motif-containing protein 5, Ubiquitin-conjugating enzyme E2 N, ... | | Authors: | Johnson, R, Taylor, A.B, Hart, P.J, Ivanov, D.N. | | Deposit date: | 2014-05-27 | | Release date: | 2015-07-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | RING Dimerization Links Higher-Order Assembly of TRIM5 alpha to Synthesis of K63-Linked Polyubiquitin.
Cell Rep, 12, 2015
|
|
4NV0
 
 | | Crystal structure of cytosolic 5'-nucleotidase IIIB (cN-IIIB) bound to 7-methylguanosine | | Descriptor: | 7-METHYLGUANOSINE, 7-methylguanosine phosphate-specific 5'-nucleotidase, MAGNESIUM ION, ... | | Authors: | Monecke, T, Neumann, P, Ficner, R. | | Deposit date: | 2013-12-04 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structures of the Novel Cytosolic 5'-Nucleotidase IIIB Explain Its Preference for m7GMP
Plos One, 9, 2014
|
|
6ZX7
 
 | |
6VKZ
 
 | |
3J6U
 
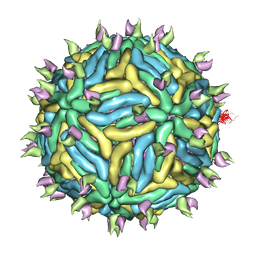 | | Cryo-EM structure of Dengue virus serotype 3 in complex with human antibody 5J7 Fab | | Descriptor: | Fab 5J7 heavy chain, Fab 5J7 light chain, envelope protein, ... | | Authors: | Fibriansah, G, Tan, J.L, Smith, S.A, de Alwis, R, Ng, T.-S, Kostyuchenko, V.A, Kukkaro, P, de Silva, A.M, Crowe Jr, J.E, Lok, S.-M. | | Deposit date: | 2014-03-26 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | A highly potent human antibody neutralizes dengue virus serotype 3 by binding across three surface proteins.
Nat Commun, 6, 2015
|
|
3J9P
 
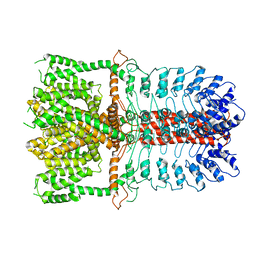 | | Structure of the TRPA1 ion channel determined by electron cryo-microscopy | | Descriptor: | Maltose-binding periplasmic protein, Transient receptor potential cation channel subfamily A member 1 chimera | | Authors: | Paulsen, C.E, Armache, J.-P, Gao, Y, Cheng, Y, Julius, D. | | Deposit date: | 2015-02-14 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.24 Å) | | Cite: | Structure of the TRPA1 ion channel suggests regulatory mechanisms.
Nature, 520, 2015
|
|
4NYQ
 
 | | In-vivo crystallisation (midguts of a viviparous cockroach) and structure at 1.2 A resolution of a glycosylated, lipid-binding, lipocalin-like protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Coussens, N.P, Gallat, F.-X, Ramaswamy, S, Yagi, K, Tobe, S.S, Stay, B, Chavas, L.M.G. | | Deposit date: | 2013-12-11 | | Release date: | 2014-01-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of a heterogeneous, glycosylated, lipid-bound, in vivo-grown protein crystal at atomic resolution from the viviparous cockroach Diploptera punctata.
Iucrj, 3, 2016
|
|
4TQF
 
 | | Crystal Structure of the C-terminal domain of IFRS bound with 2-(5-bromothienyl)-L-Ala and ATP | | Descriptor: | 1,2-ETHANEDIOL, 3-(5-bromothiophen-2-yl)-L-alanine, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Nakamura, A, O'Donoghue, P, Soll, D. | | Deposit date: | 2014-06-11 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7143 Å) | | Cite: | Polyspecific pyrrolysyl-tRNA synthetases from directed evolution.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4NZV
 
 | | DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities | | Descriptor: | Exonuclease, putative, MANGANESE (II) ION | | Authors: | Shibata, A, Moiani, D, Arvai, A.S, Perry, J, Harding, S.M, Genois, M, Maity, R, Rossum-Fikkert, S, Kertokalio, A, Romoli, F, Ismail, A, Ismalaj, E, Petricci, E, Neale, M.J, Bristow, R.G, Masson, J, Wyman, C, Jeggo, P.A, Tainer, J.A. | | Deposit date: | 2013-12-12 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities.
Mol.Cell, 53, 2014
|
|
7AQ8
 
 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H583Y/D576A | | Descriptor: | (MU-4-SULFIDO)-TETRA-NUCLEAR COPPER ION, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Zhang, L, Bill, E, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-10-20 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.791 Å) | | Cite: | Histidine-Gated Proton-Coupled Electron Transfer to the Cu A Site of Nitrous Oxide Reductase.
J.Am.Chem.Soc., 143, 2021
|
|
1LC2
 
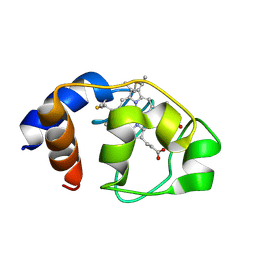 | |
4TR6
 
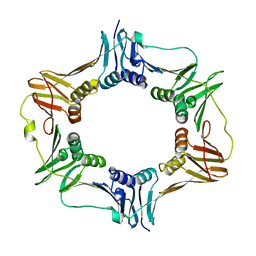 | | Crystal structure of DNA polymerase sliding clamp from Bacillus subtilis | | Descriptor: | DNA polymerase III subunit beta, SODIUM ION | | Authors: | Burnouf, D, Olieric, V, Ennifar, E, Wolff, P. | | Deposit date: | 2014-06-14 | | Release date: | 2014-09-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Differential Modes of Peptide Binding onto Replicative Sliding Clamps from Various Bacterial Origins.
J.Med.Chem., 57, 2014
|
|
3K1K
 
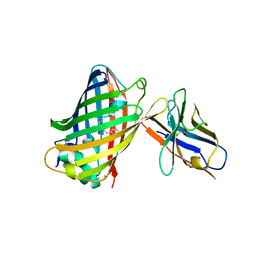 | | Green fluorescent protein bound to enhancer nanobody | | Descriptor: | Enhancer, Green Fluorescent Protein | | Authors: | Kirchhofer, A, Helma, J, Schmidthals, K, Frauer, C, Cui, S, Karcher, A, Pellis, M, Muyldermans, S, Delucci, C.C, Cardoso, M.C, Leonhardt, H, Hopfner, K.-P, Rothbauer, U. | | Deposit date: | 2009-09-28 | | Release date: | 2009-12-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Modulation of protein properties in living cells using nanobodies
Nat.Struct.Mol.Biol., 17, 2010
|
|
3K3J
 
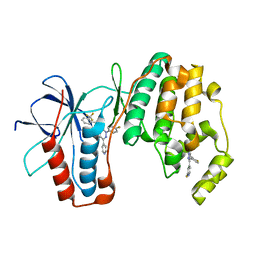 | | P38alpha bound to novel DFG-out compound PF-00416121 | | Descriptor: | 2-(4-fluorophenyl)-3-oxo-6-pyridin-4-yl-N-[2-(trifluoromethyl)benzyl]-2,3-dihydropyridazine-4-carboxamide, 2-fluoro-4-[4-(4-fluorophenyl)-1H-pyrazol-3-yl]pyridine, Mitogen-activated protein kinase 14 | | Authors: | Kazmirski, S.L, DiNitto, J.P. | | Deposit date: | 2009-10-02 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | The Design, Synthesis and Potential Utility of Fluorescence Probes that Target DFG-out Conformation of p38alpha for High Throughput Screening Binding Assay.
Chem.Biol.Drug Des., 74, 2009
|
|
1LKQ
 
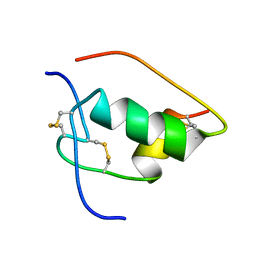 | | NMR STRUCTURE OF HUMAN INSULIN MUTANT ILE-A2-GLY, VAL-A3-GLY, HIS-B10-ASP, PRO-B28-LYS, LYS-B29-PRO, 20 STRUCTURES | | Descriptor: | INSULIN | | Authors: | Weiss, M.A, Hua, Q.X, Chu, Y.C, Jia, W, Philips, N.F, Wang, R.Y, Katsoyannis, P.G. | | Deposit date: | 2002-04-25 | | Release date: | 2002-05-22 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Mechanism of insulin chain combination. Asymmetric roles of A-chain alpha-helices in disulfide pairing
J.Biol.Chem., 277, 2002
|
|
1L5J
 
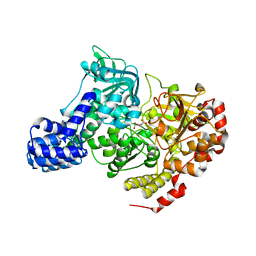 | | CRYSTAL STRUCTURE OF E. COLI ACONITASE B. | | Descriptor: | ACONITATE ION, Aconitate hydratase 2, FE3-S4 CLUSTER | | Authors: | Williams, C.H, Stillman, T.J, Barynin, V.V, Sedelnikova, S.E, Tang, Y, Green, J, Guest, J.R, Artymiuk, P.J. | | Deposit date: | 2002-03-07 | | Release date: | 2002-06-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | E. coli aconitase B structure reveals a HEAT-like domain with implications for protein-protein recognition.
Nat.Struct.Biol., 9, 2002
|
|
4NFE
 
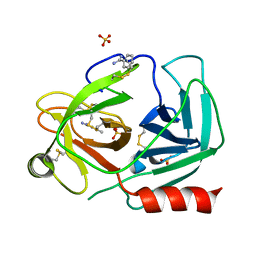 | | Human kallikrein-related peptidase 2 in complex with benzamidine | | Descriptor: | BENZAMIDINE, Kallikrein-2, SULFATE ION | | Authors: | Skala, W, Brandstetter, H, Magdolen, V, Goettig, P. | | Deposit date: | 2013-10-31 | | Release date: | 2014-10-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-function analyses of human kallikrein-related peptidase 2 establish the 99-loop as master regulator of activity
J.Biol.Chem., 289, 2014
|
|
1LN6
 
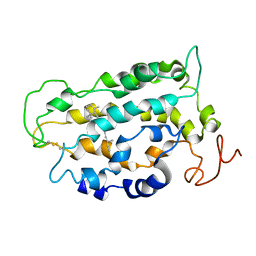 | | STRUCTURE OF BOVINE RHODOPSIN (Metarhodopsin II) | | Descriptor: | RETINAL, RHODOPSIN | | Authors: | Choi, G, Landin, J, Galan, J.F, Birge, R.R, Albert, A.D, Yeagle, P.L. | | Deposit date: | 2002-05-03 | | Release date: | 2002-07-10 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structural studies of metarhodopsin II, the activated form of the G-protein coupled receptor, rhodopsin.
Biochemistry, 41, 2002
|
|
