6YOG
 
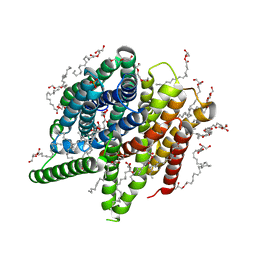 | | Structure of PepTSt from COC IMISX setup collected by still serial crystallography on crystals prelocated by 2D X-ray phase-contrast imaging | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, 2-(2-METHOXYETHOXY)ETHANOL, Di-or tripeptide:H+ symporter, ... | | Authors: | Huang, C.-Y, Martiel, I, Villanueva-Perez, P, Panepucci, E, Caffrey, M, Wang, M. | | Deposit date: | 2020-04-14 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Low-dose in situ prelocation of protein microcrystals by 2D X-ray phase-contrast imaging for serial crystallography.
Iucrj, 7, 2020
|
|
9BKF
 
 | |
6T6D
 
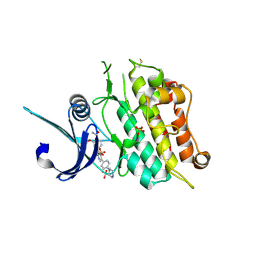 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound M4K2149 | | Descriptor: | 2-methoxy-4-[4-methyl-5-(4-piperazin-1-ylphenyl)pyridin-3-yl]benzamide, Activin receptor type I, SULFATE ION | | Authors: | Adamson, R.J, Williams, E.P, Smil, D, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2019-10-18 | | Release date: | 2019-10-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Targeting ALK2: An Open Science Approach to Developing Therapeutics for the Treatment of Diffuse Intrinsic Pontine Glioma.
J.Med.Chem., 63, 2020
|
|
8WI1
 
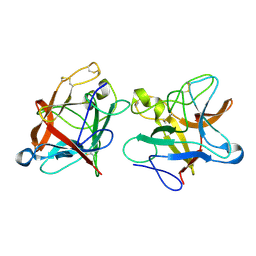 | |
6KK5
 
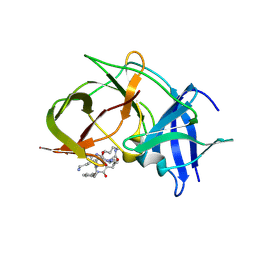 | | Crystal structure of Zika NS2B-NS3 protease with compound 15 | | Descriptor: | 1-[(5~{S},8~{R},15~{S},18~{S})-15,18-bis(4-azanylbutyl)-5-methyl-4,7,14,17,20-pentakis(oxidanylidene)-3,6,13,16,19-pentazabicyclo[20.3.1]hexacosa-1(25),22(26),23-trien-8-yl]guanidine, NS3 protease, Serine protease subunit NS2B | | Authors: | Quek, J.P. | | Deposit date: | 2019-07-23 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structure-Based Macrocyclization of Substrate Analogue NS2B-NS3 Protease Inhibitors of Zika, West Nile and Dengue viruses.
Chemmedchem, 15, 2020
|
|
8Z9Z
 
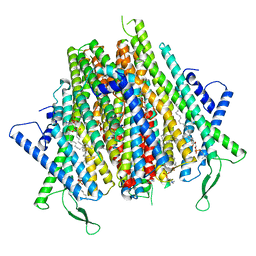 | | Cryo-EM structure of the insect olfactory receptor OR5-Orco heterocomplex from Acyrthosiphon pisum | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Odorant receptor, ApisOR5, ... | | Authors: | Wang, Y.D, Qiu, L, Guan, Z.Y, Wang, Q, Wang, G.R, Yin, P. | | Deposit date: | 2024-04-24 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for odorant recognition of the insect odorant receptor OR-Orco heterocomplex.
Science, 384, 2024
|
|
6GUC
 
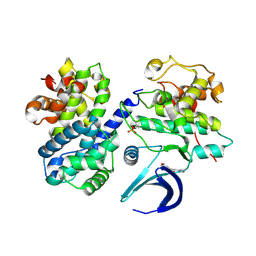 | | CDK2/CyclinA in complex with SU9516 | | Descriptor: | (3Z)-3-(1H-IMIDAZOL-5-YLMETHYLENE)-5-METHOXY-1H-INDOL-2(3H)-ONE, Cyclin-A2, Cyclin-dependent kinase 2 | | Authors: | Wood, D.J, Korolchuk, S, Tatum, N.J, Wang, L.Z, Endicott, J.A, Noble, M.E.M, Martin, M.P. | | Deposit date: | 2018-06-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition.
Cell Chem Biol, 26, 2019
|
|
8VOL
 
 | |
4KVA
 
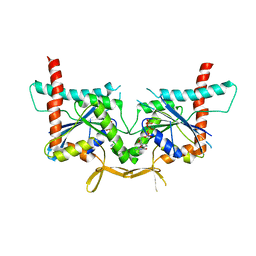 | | GTPase domain of Septin 10 from Schistosoma mansoni in complex with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Septin | | Authors: | Zeraik, A.E, Pereira, H.M, Santos, Y.V, Brandao-Neto, J, Garratt, R.C, Araujo, A.P.U, Demarco, R. | | Deposit date: | 2013-05-22 | | Release date: | 2014-02-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal Structure of a Schistosoma mansoni Septin Reveals the Phenomenon of Strand Slippage in Septins Dependent on the Nature of the Bound Nucleotide.
J.Biol.Chem., 289, 2014
|
|
6GRP
 
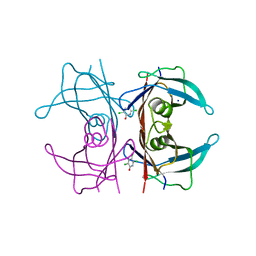 | | Crystal Structure Of Human Transthyretin in complex with 3,5,6-trichloro-2-pyridinol (TC2P) | | Descriptor: | 3,5,6-trichloro-2-pyridinol, SODIUM ION, Transthyretin | | Authors: | Grundstrom, C, Zhang, J, Olofsson, A, Andersson, P.L, Sauer-Eriksson, A.E. | | Deposit date: | 2018-06-12 | | Release date: | 2018-07-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Interspecies Variation between Fish and Human Transthyretins in Their Binding of Thyroid-Disrupting Chemicals.
Environ. Sci. Technol., 52, 2018
|
|
8YVW
 
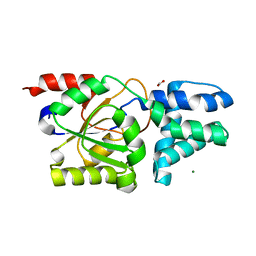 | | Crystal structure of D12N mutant of L-azetidine-2-carboxylate hydrolase | | Descriptor: | (S)-2-haloacid dehalogenase, FORMIC ACID, IMIDAZOLE, ... | | Authors: | Toyoda, M, Mizutani, K, Mikami, B, Wackett, L.P, Esaki, N, Kurihara, T. | | Deposit date: | 2024-03-29 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Research for the crystal structure of L-azetidine-2-carboxylate hydrolase
To Be Published
|
|
6GEZ
 
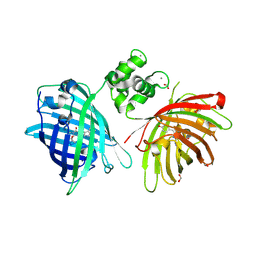 | | THE STRUCTURE OF TWITCH-2B N532F | | Descriptor: | CALCIUM ION, FORMIC ACID, Green fluorescent protein,Optimized Ratiometric Calcium Sensor,Green fluorescent protein,Green fluorescent protein | | Authors: | Trigo Mourino, P, Paulat, M, Thestrup, T, Griesbeck, O, Griesinger, C, Becker, S. | | Deposit date: | 2018-04-27 | | Release date: | 2019-08-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Dynamic tuning of FRET in a green fluorescent protein biosensor.
Sci Adv, 5, 2019
|
|
8YSA
 
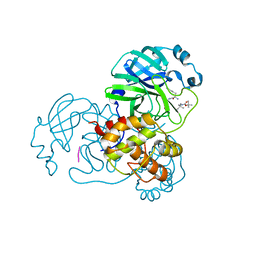 | | The co-crystal structure of SARS-CoV-2 Mpro in complex with compound H102 | | Descriptor: | 3C-like proteinase nsp5, BOC-TBG-PHE-ELL | | Authors: | Zheng, W.Y, Fu, L.F, Feng, Y, Han, P, Qi, J.X. | | Deposit date: | 2024-03-22 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery, Biological Activity, and Structural Mechanism of a Potent Inhibitor of SARS-CoV-2 Main Protease
To Be Published
|
|
8WIH
 
 | | Crystal structure of E. coli ThrS catalytic domain mutant G463A in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Threonine--tRNA ligase, ZINC ION | | Authors: | Qiao, H, Wang, Z, Wang, J, Fang, P. | | Deposit date: | 2023-09-24 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Specific glycine-dependent enzyme motion determines the potency of conformation selective inhibitors of threonyl-tRNA synthetase.
Commun Biol, 7, 2024
|
|
9IK2
 
 | | The co-crystal structure of SARS-CoV-2 Mpro in complex with compound H109 | | Descriptor: | 3C-like proteinase, tert-butyl N-[(2S)-1-[[(2S)-1-[[(2S)-1-azanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-1-oxidanylidene-3-phenyl-propan-2-yl]amino]-3,3-dimethyl-1-oxidanylidene-butan-2-yl]carbamate | | Authors: | Feng, Y, Zheng, W.Y, Han, P, Fu, L.F, Qi, J.X. | | Deposit date: | 2024-06-26 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-guided discovery of a small molecule inhibitor of SARS-CoV-2 main protease with potent in vitro and in vivo antiviral activities
To Be Published
|
|
8Z9U
 
 | |
8VT8
 
 | | UIC-13-BIF-A4Dab NBD-Cl binding | | Descriptor: | 4-chloro-7-nitrobenzofurazan, ACETONITRILE, UIC-13-BIF-A4Dab-NBD-Cl | | Authors: | Ganatra, P. | | Deposit date: | 2024-01-26 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Diverse Proteomimetic Frameworks via Rational Design of pi-Stacking Peptide Tectons.
J.Am.Chem.Soc., 146, 2024
|
|
8W1V
 
 | | The beta2 adrenergic receptor bound to a bitopic ligand | | Descriptor: | (2S)-1-[(3-{1-[4-(4-{(2S)-2-hydroxy-3-[(propan-2-yl)amino]propoxy}phenyl)butyl]-1H-1,2,3-triazol-4-yl}propyl)amino]-3-(2-propylphenoxy)propan-2-ol, Beta-2 adrenergic receptor,Endolysin, Lauryl Maltose Neopentyl Glycol, ... | | Authors: | Gaiser, B, Danielsen, M, Xu, X, Jorgensen, K, Fronik, P, Marcher-Rorsted, E, Wrobe, T, Hirata, K, Liu, X, Mathiesen, J, Pedersen, D. | | Deposit date: | 2024-02-19 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Bitopic Ligands Support the Presence of a Metastable Binding Site at the beta 2 Adrenergic Receptor.
J.Med.Chem., 67, 2024
|
|
6GOO
 
 | | Crystal Structure Of Sea Bream Transthyretin in complex with Perfluorooctanoic acid (PFOA). Crystallized in AmSO4 | | Descriptor: | Transthyretin, pentadecafluorooctanoic acid | | Authors: | Grundstrom, C, Zhang, J, Olofsson, A, Andersson, P.L, Sauer-Eriksson, A.E. | | Deposit date: | 2018-06-01 | | Release date: | 2018-07-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Interspecies Variation between Fish and Human Transthyretins in Their Binding of Thyroid-Disrupting Chemicals.
Environ. Sci. Technol., 52, 2018
|
|
9BKP
 
 | |
7NSR
 
 | | Myelin protein P2 I50del | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Myelin P2 protein, ... | | Authors: | Uusitalo, M, Ruskamo, S, Kursula, P. | | Deposit date: | 2021-03-08 | | Release date: | 2021-09-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Human myelin protein P2: from crystallography to time-lapse membrane imaging and neuropathy-associated variants.
Febs J., 288, 2021
|
|
9BOK
 
 | | Crystal structure of reduced bovine trypsin, 50mM DTT-treated | | Descriptor: | BENZAMIDINE, Cationic trypsin | | Authors: | Zhou, D, Chen, L, Rose, J.P, Wang, B.C. | | Deposit date: | 2024-05-03 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of reduced bovine trypsin, 50mM DTT-treated
To Be Published
|
|
8X2W
 
 | |
4O8P
 
 | | Crystal structure of SthAraf62A, a GH62 family alpha-L-arabinofuranosidase from Streptomyces thermoviolaceus, bound to xylotetraose | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, Alpha-L-arabinofuranosidase, CALCIUM ION, ... | | Authors: | Stogios, P.J, Wang, W, Xu, X, Cui, H, Master, E, Savchenko, A. | | Deposit date: | 2013-12-28 | | Release date: | 2014-07-02 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (1.557 Å) | | Cite: | Elucidation of the molecular basis for arabinoxylan-debranching activity of a thermostable family GH62 alpha-l-arabinofuranosidase from Streptomyces thermoviolaceus.
Appl.Environ.Microbiol., 80, 2014
|
|
9FCU
 
 | | CysG(N-16)-H121A mutant in complex with SAH from Kitasatospora cystarginea | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SAM-dependent methyltransferase | | Authors: | Kuttenlochner, W, Beller, P, Kaysser, L, Groll, M. | | Deposit date: | 2024-05-15 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Deciphering the SAM- and metal-dependent mechanism of O-methyltransferases in cystargolide and belactosin biosynthesis: A structure-activity relationship study.
J.Biol.Chem., 300, 2024
|
|
