8FNU
 
 | | Structure of RdrA from Streptococcus suis RADAR defense system | | Descriptor: | KAP NTPase domain-containing protein | | Authors: | Duncan-Lowey, B, Johnson, A.G, Rawson, S, Mayer, M.L, Kranzusch, P.J. | | Deposit date: | 2022-12-28 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structure of the RADAR supramolecular anti-phage defense complex.
Cell, 186, 2023
|
|
5F2E
 
 | | Crystal Structure of small molecule ARS-853 covalently bound to K-Ras G12C | | Descriptor: | 1-[3-[4-[2-[[4-chloranyl-5-(1-methylcyclopropyl)-2-oxidanyl-phenyl]amino]ethanoyl]piperazin-1-yl]azetidin-1-yl]prop-2-en-1-one, GLYCEROL, GLYCINE, ... | | Authors: | Patricelli, M.P, Janes, M.R, Li, L.-S, Hansen, R, Peters, U, Kessler, L.V, Chen, Y, Kucharski, J.M, Feng, J, Ely, T, Chen, J.H, Firdaus, S.J, Babbar, A, Ren, P, Liu, Y. | | Deposit date: | 2015-12-01 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Selective Inhibition of Oncogenic KRAS Output with Small Molecules Targeting the Inactive State.
Cancer Discov, 6, 2016
|
|
6X6Q
 
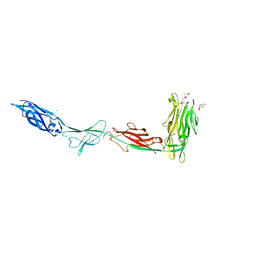 | |
5YGA
 
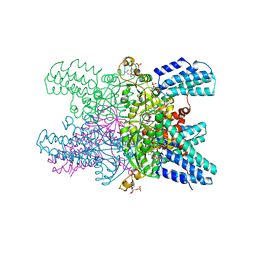 | | Crystal structure of ribose-1,5-bisphosphate isomerase mutant D204N from Pyrococcus horikoshii OT3 in complex with ribose-1,5-bisphosphate, AMP and GMP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,5-di-O-phosphono-alpha-D-ribofuranose, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Gogoi, P, Kanaujia, S.P. | | Deposit date: | 2017-09-22 | | Release date: | 2018-02-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A presumed homologue of the regulatory subunits of eIF2B functions as ribose-1,5-bisphosphate isomerase in Pyrococcus horikoshii OT3.
Sci Rep, 8, 2018
|
|
6X6M
 
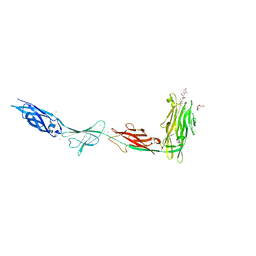 | |
6XEU
 
 | | CryoEM structure of GIRK2PIP2* - G protein-gated inwardly rectifying potassium channel GIRK2 with PIP2 | | Descriptor: | G protein-activated inward rectifier potassium channel 2, POTASSIUM ION, SODIUM ION, ... | | Authors: | Mathiharan, Y.K, Glaaser, I.W, Skiniotis, G, Slesinger, P.A. | | Deposit date: | 2020-06-13 | | Release date: | 2021-09-01 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into GIRK2 channel modulation by cholesterol and PIP2
Cell Rep, 36, 2021
|
|
6XEV
 
 | | CryoEM structure of GIRK2-PIP2/CHS - G protein-gated inwardly rectifying potassium channel GIRK2 with modulators cholesteryl hemisuccinate and PIP2 | | Descriptor: | CHOLESTEROL HEMISUCCINATE, G protein-activated inward rectifier potassium channel 2, POTASSIUM ION, ... | | Authors: | Mathiharan, Y.K, Glaaser, I.W, Skiniotis, G, Slesinger, P.A. | | Deposit date: | 2020-06-14 | | Release date: | 2021-09-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into GIRK2 channel modulation by cholesterol and PIP2
Cell Rep, 36, 2021
|
|
4QEM
 
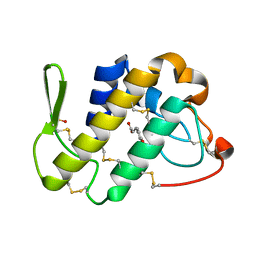 | | Crystal structure of the complex of Phospholipase A2 With P-Coumaric Acid At 1.2 A Resolution | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Phospholipase A2 VRV-PL-VIIIa, SULFATE ION | | Authors: | Shukla, P.K, Tiwari, P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-05-17 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structures and binding studies of the complexes of phospholipase A2 with five inhibitors
Biochim.Biophys.Acta, 1854, 2015
|
|
8FGW
 
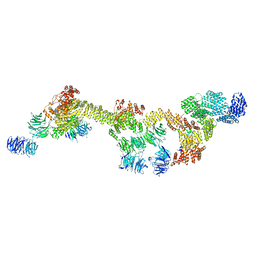 | | Human IFT-A complex structures provide molecular insights into ciliary transport | | Descriptor: | Intraflagellar transport protein 122 homolog, Intraflagellar transport protein 140 homolog, Intraflagellar transport protein 43 homolog, ... | | Authors: | Jiang, M, Palicharla, V.R, Miller, D, Hwang, S.H, Zhu, H, Hixson, P, Mukhopadhyay, S, Sun, J. | | Deposit date: | 2022-12-12 | | Release date: | 2023-02-22 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Human IFT-A complex structures provide molecular insights into ciliary transport.
Cell Res., 33, 2023
|
|
8FH3
 
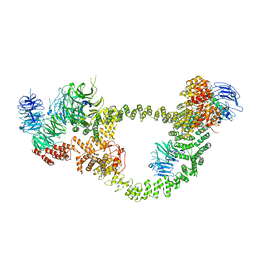 | | Human IFT-A complex structures provide molecular insights into ciliary transport | | Descriptor: | Intraflagellar transport protein 122 homolog, Intraflagellar transport protein 140 homolog, Tubby-related protein 3, ... | | Authors: | Jiang, M, Palicharla, V.R, Miller, D, Hwang, S.H, Zhu, H, Hixson, P, Mukhopadhyay, S, Sun, J. | | Deposit date: | 2022-12-13 | | Release date: | 2023-02-22 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Human IFT-A complex structures provide molecular insights into ciliary transport.
Cell Res., 33, 2023
|
|
7SGF
 
 | | Lassa virus glycoprotein construct (Josiah GPC-I53-50A) in complex with LAVA01 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Antanasijevic, A, Brouwer, P.J.M, Ward, A.B. | | Deposit date: | 2021-10-05 | | Release date: | 2022-10-12 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (4.41 Å) | | Cite: | Lassa virus glycoprotein nanoparticles elicit neutralizing antibody responses and protection.
Cell Host Microbe, 30, 2022
|
|
5YL0
 
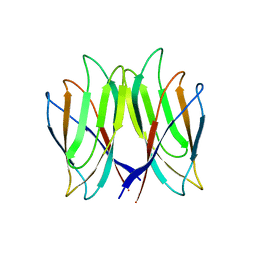 | | The crystal structure of Penaeus vannamei nodavirus P-domain (P212121) | | Descriptor: | Capsid protein | | Authors: | Chen, N.C, Yoshimura, M, Lin, C.C, Guan, H.H, Chuankhayan, P, Chen, C.J. | | Deposit date: | 2017-10-16 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | The atomic structures of shrimp nodaviruses reveal new dimeric spike structures and particle polymorphism.
Commun Biol, 2, 2019
|
|
5YLF
 
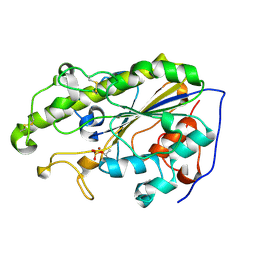 | | MCR-1 complex with D-glucose | | Descriptor: | Probable phosphatidylethanolamine transferase Mcr-1, ZINC ION, beta-D-glucopyranose | | Authors: | Wei, P.C, Song, G.J, Shi, M.Y, Zhou, Y.F, Liu, Y, Lei, J, Chen, P, Yin, L. | | Deposit date: | 2017-10-17 | | Release date: | 2017-11-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Substrate analog interaction with MCR-1 offers insight into the rising threat of the plasmid-mediated transferable colistin resistance.
FASEB J., 32, 2018
|
|
8FPT
 
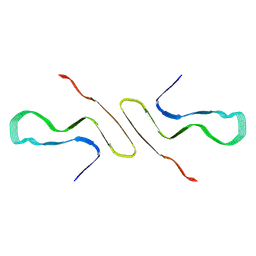 | | STRUCTURE OF ALPHA-SYNUCLEIN FIBRILS DERIVED FROM HUMAN LEWY BODY DEMENTIA TISSUE | | Descriptor: | Alpha-synuclein | | Authors: | Barclay, A.M, Dhavale, D.D, Borcik, C.G, Rau, M.J, Basore, K, Milchberg, M.H, Warmuth, O.A, Kotzbauer, P.T, Rienstra, C.M, Schwieters, C.D. | | Deposit date: | 2023-01-05 | | Release date: | 2023-02-22 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Structure of alpha-synuclein fibrils derived from human Lewy body dementia tissue.
Biorxiv, 2023
|
|
6S0B
 
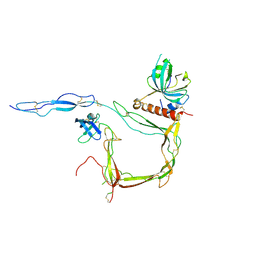 | | Crystal Structure of Properdin in complex with the CTC domain of C3/C3b | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C3, Properdin, ... | | Authors: | van den Bos, R.M, Pearce, N.M, Gros, P. | | Deposit date: | 2019-06-14 | | Release date: | 2019-09-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.312 Å) | | Cite: | Insights Into Enhanced Complement Activation by Structures of Properdin and Its Complex With the C-Terminal Domain of C3b.
Front Immunol, 10, 2019
|
|
8TY1
 
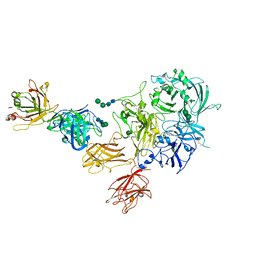 | |
8P51
 
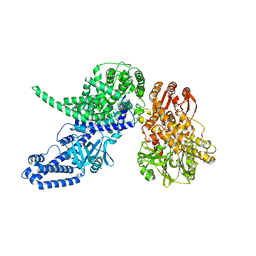 | |
6M6Z
 
 | | A de novo designed transmembrane nanopore, TMH4C4 | | Descriptor: | TMH4C4 | | Authors: | Lu, P, Xu, C, Reggiano, G, Xu, Q, DiMaio, F, Baker, D. | | Deposit date: | 2020-03-16 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Computational design of transmembrane pores.
Nature, 585, 2020
|
|
8P50
 
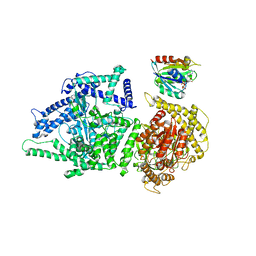 | | Photorhabdus luminescens Makes caterpillars floppy (Mcf) toxin with the C-terminal deletion in complex with Arf3 | | Descriptor: | ADP-ribosylation factor 3, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Belyy, A, Heilen, P, Hofnagel, O, Raunser, S. | | Deposit date: | 2023-05-23 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | Structure and activation mechanism of the Makes caterpillars floppy 1 toxin.
Nat Commun, 14, 2023
|
|
9BK3
 
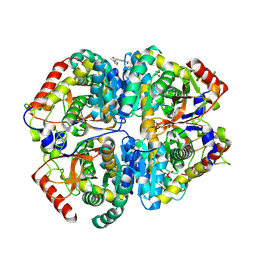 | | Crystal structure of Lactate dehydrogenase in complex with 4-((4-(1-methyl-1H-imidazole-2-carbonyl)phenyl)amino)-4-oxo-2-(4-(trifluoromethyl)phenyl)butanoic acid (R-enantiomer, orthorhombic P form) | | Descriptor: | (2R)-4-[4-(1-methyl-1H-imidazole-2-carbonyl)anilino]-4-oxo-2-[4-(trifluoromethyl)phenyl]butanoic acid, CHLORIDE ION, L-lactate dehydrogenase A chain, ... | | Authors: | Lovell, S, Cooper, A, Battaile, K.P, Sharma, H. | | Deposit date: | 2024-04-26 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthesis and biological characterization of an orally bioavailable lactate dehydrogenase-A inhibitor against pancreatic cancer.
Eur.J.Med.Chem., 275, 2024
|
|
7YWL
 
 | | Six DNA Helix Bundle nanopore - State 3 | | Descriptor: | DNA (50-MER) | | Authors: | Javed, A, Ahmad, K, Lanphere, C, Coveney, P, Howorka, S, Orlova, E.V. | | Deposit date: | 2022-02-14 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structure and dynamics of an archetypal DNA nanoarchitecture revealed via cryo-EM and molecular dynamics simulations.
Nat Commun, 14, 2023
|
|
7YWI
 
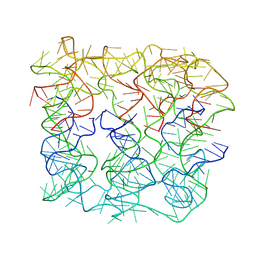 | | Six DNA duplex bundle nanopore - State 2 | | Descriptor: | DNA (50-MER) | | Authors: | Javed, A, Ahmad, K, Lanphere, C, Coveney, P, Howorka, S, Orlova, E.V. | | Deposit date: | 2022-02-14 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structure and dynamics of an archetypal DNA nanoarchitecture revealed via cryo-EM and molecular dynamics simulations.
Nat Commun, 14, 2023
|
|
6LRZ
 
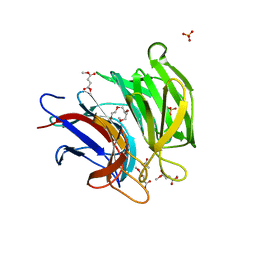 | | Crystal structure of Keap1 in complex with dimethyl fumarate (DMF) | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, ACETATE ION, Keap1-DC, ... | | Authors: | Padmanabhan, B, Unni, S, Deshmukh, P. | | Deposit date: | 2020-01-16 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural insights into the multiple binding modes of Dimethyl Fumarate (DMF) and its analogs to the Kelch domain of Keap1.
Febs J., 288, 2021
|
|
7YWO
 
 | | Six DNA Helix Bundle nanopore - State 5 | | Descriptor: | DNA (50-MER) | | Authors: | Javed, A, Ahmad, K, Lanphere, C, Coveney, P, Howorka, S, Orlova, E.V. | | Deposit date: | 2022-02-14 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structure and dynamics of an archetypal DNA nanoarchitecture revealed via cryo-EM and molecular dynamics simulations.
Nat Commun, 14, 2023
|
|
7YWN
 
 | | Six DNA Helix Bundle nanopore - State 4 | | Descriptor: | DNA (50-MER) | | Authors: | Javed, A, Ahmad, K, Lanphere, C, Orlova, E.V, Coveney, P, Howorka, S. | | Deposit date: | 2022-02-14 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structure and dynamics of an archetypal DNA nanoarchitecture revealed via cryo-EM and molecular dynamics simulations.
Nat Commun, 14, 2023
|
|
