7ZSB
 
 | |
7BGP
 
 | | Crystal structure of MG-132 covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 in absence of DTT. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | Deposit date: | 2021-01-08 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
7C0V
 
 | | Crystal structure of a dinucleotide-binding protein (Y224A) of ABC transporter endogenously bound to uridylyl-3'-5'-phospho-guanosine (Form I) | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, CARBON DIOXIDE, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-01 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and thermodynamic insights into the novel dinucleotide-binding protein of ABC transporter unveils its moonlighting function.
Febs J., 288, 2021
|
|
7C15
 
 | | Crystal structure of a dinucleotide-binding protein (Q274A) of ABC transporter endogenously bound to uridylyl-3'-5'-phospho-guanosine (Form II) | | Descriptor: | 1,2-ETHANEDIOL, CARBON DIOXIDE, CARBONATE ION, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-02 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and thermodynamic insights into the novel dinucleotide-binding protein of ABC transporter unveils its moonlighting function.
Febs J., 288, 2021
|
|
7C16
 
 | | Crystal structure of a dinucleotide-binding protein (Y224A/Y246A) of ABC transporter endogenously bound to uridylyl-3'-5'-phospho-guanosine (Form I) | | Descriptor: | 1,2-ETHANEDIOL, CARBON DIOXIDE, CARBONATE ION, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-02 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and thermodynamic insights into the novel dinucleotide-binding protein of ABC transporter unveils its moonlighting function.
Febs J., 288, 2021
|
|
7BB2
 
 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 at 1.6A resolution (spacegroup P2(1)2(1)2(1)) | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | Deposit date: | 2020-12-16 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
7C0L
 
 | |
7C0U
 
 | | Crystal structure of a dinucleotide-binding protein (S127A) of ABC transporter endogenously bound to uridylyl-3'-5'-phospho-guanosine | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-01 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and thermodynamic insights into the novel dinucleotide-binding protein of ABC transporter unveils its moonlighting function.
Febs J., 288, 2021
|
|
7O45
 
 | | Crystal structure of ADD domain of the human DNMT3B methyltransferase | | Descriptor: | BROMIDE ION, Isoform 6 of DNA (cytosine-5)-methyltransferase 3B, ZINC ION | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Bonchuk, A.N, Georgiev, P.G, Popov, V.O. | | Deposit date: | 2021-04-05 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the DNMT3B ADD domain suggests the absence of a DNMT3A-like autoinhibitory mechanism.
Biochem.Biophys.Res.Commun., 619, 2022
|
|
7C0O
 
 | | Crystal structure of a dinucleotide-binding protein (Y56F) of ABC transporter endogenously bound to uridylyl-3'-5'-phospho-guanosine | | Descriptor: | 1,2-ETHANEDIOL, CARBON DIOXIDE, CARBONATE ION, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-01 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and thermodynamic insights into the novel dinucleotide-binding protein of ABC transporter unveils its moonlighting function.
Febs J., 288, 2021
|
|
5ZC2
 
 | | Acinetobacter baumannii p-hydroxyphenylacetate 3-hydroxylase (HPAH), reductase component (C1) | | Descriptor: | FLAVIN MONONUCLEOTIDE, p-hydroxyphenylacetate 3-hydroxylase, reductase component | | Authors: | Yuenyao, A, Petchyam, N, Chaiyen, P, Pakotiprapha, D. | | Deposit date: | 2018-02-14 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.898 Å) | | Cite: | Crystal structure of the flavin reductase of Acinetobacter baumannii p-hydroxyphenylacetate 3-hydroxylase (HPAH) and identification of amino acid residues underlying its regulation by aromatic ligands
Arch. Biochem. Biophys., 653, 2018
|
|
7C0R
 
 | | Crystal structure of a dinucleotide-binding protein (F79A) of ABC transporter endogenously bound to uridylyl-3'-5'-phospho-guanosine (Form I) | | Descriptor: | 1,2-ETHANEDIOL, CARBON DIOXIDE, CHLORIDE ION, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-01 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and thermodynamic insights into the novel dinucleotide-binding protein of ABC transporter unveils its moonlighting function.
Febs J., 288, 2021
|
|
6RZ9
 
 | | Crystal structure of the human cysteinyl leukotriene receptor 2 in complex with ONO-2770372 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2~{S})-8-[[4-[4-(5-fluoranyl-2-methyl-phenyl)butoxy]phenyl]carbonylamino]-4-(4-oxidanyl-4-oxidanylidene-butyl)-2,3-dih ydro-1,4-benzoxazine-2-carboxylic acid, CHOLESTEROL, ... | | Authors: | Gusach, A, Luginina, A, Marin, E, Brouillette, R.L, Besserer-Offroy, E, Longpre, J.M, Ishchenko, A, Popov, P, Fujimoto, T, Maruyama, T, Stauch, B, Ergasheva, M, Romanovskaya, D, Stepko, A, Kovalev, K, Shevtsov, M, Gordeliy, V, Han, G.W, Sarret, P, Katritch, V, Borshchevskiy, V, Mishin, A, Cherezov, V. | | Deposit date: | 2019-06-12 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structural basis of ligand selectivity and disease mutations in cysteinyl leukotriene receptors.
Nat Commun, 10, 2019
|
|
8CMF
 
 | | Human Leukocyte Antigen class II allotype DR1 presenting SARS-CoV-2 nsp3 epitope (orf1ab)1350-1364 | | Descriptor: | 1,2-ETHANEDIOL, HLA class II histocompatibility antigen, DR alpha chain, ... | | Authors: | MacLachlan, B.J, Mason, G.H, Sourfield, D.O, Godkin, A.J, Rizkallah, P.J. | | Deposit date: | 2023-02-19 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural definition of HLA class II-presented SARS-CoV-2 epitopes reveals a mechanism to escape pre-existing CD4 + T cell immunity.
Cell Rep, 42, 2023
|
|
7BAD
 
 | | Crystal structure of PAFB in complex with p-sulfonato-calix[8]arene and zinc | | Descriptor: | Antifungal protein, PHOSPHATE ION, ZINC ION, ... | | Authors: | Guagnini, F, Huber, A, Alex, J.M, Marx, F, Crowley, P.B. | | Deposit date: | 2020-12-15 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Porous assembly of an antifungal protein mediated by zinc and sulfonato-calix[8]arene.
J.Struct.Biol., 213, 2021
|
|
8CMI
 
 | | Human Leukocyte Antigen class II allotype DR1 presenting SARS-CoV-2 Omicron (BA.1) Spike peptide S761-775 | | Descriptor: | 1,2-ETHANEDIOL, HLA class II histocompatibility antigen, DR alpha chain, ... | | Authors: | MacLachlan, B.J, Mason, G.H, Sourfield, D.O, Godkin, A.J, Rizkallah, P.J. | | Deposit date: | 2023-02-19 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural definition of HLA class II-presented SARS-CoV-2 epitopes reveals a mechanism to escape pre-existing CD4 + T cell immunity.
Cell Rep, 42, 2023
|
|
5NJ6
 
 | | Crystal structure of a thermostabilised human protease-activated receptor-2 (PAR2) in ternary complex with Fab3949 and AZ7188 at 4.0 angstrom resolution | | Descriptor: | Fab3949 H, Fab3949 L, Proteinase-activated receptor 2,Soluble cytochrome b562,Proteinase-activated receptor 2 | | Authors: | Cheng, R.K.Y, Fiez-Vandal, C, Schlenker, O, Edman, K, Aggeler, B, Brown, D.G, Brown, G, Cooke, R.M, Dumelin, C.E, Dore, A.S, Geschwindner, S, Grebner, C, Hermansson, N.-O, Jazayeri, A, Johansson, P, Leong, L, Prihandoko, R, Rappas, M, Soutter, H, Snijder, A, Sundstrom, L, Tehan, B, Thornton, P, Troast, D, Wiggin, G, Zhukov, A, Marshall, F.H, Dekker, N. | | Deposit date: | 2017-03-28 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural insight into allosteric modulation of protease-activated receptor 2.
Nature, 545, 2017
|
|
6VQY
 
 | | HLA-B*27:05 presenting an HIV-1 7mer peptide | | Descriptor: | 7-mer peptide, ARGININE, Beta-2-microglobulin, ... | | Authors: | Pymm, P, Tenzer, S, Wee, E, Weimershaus, M, Burgevin, A, Kollnberger, S, Gerstoft, J, Josephs, T.M, Ladell, K, Mclaren, J.E, Appay, V, Price, D.A, Fugger, L, Bell, J.I, Hansjorg, S, Van Endert, P, Harkiolaki, M, Iversen, A.K.N. | | Deposit date: | 2020-02-06 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Epitope length variants balance protective immune responses and viral escape in HIV-1 infection
Cell Rep, 38, 2022
|
|
5NHF
 
 | | Human Erk2 with an Erk1/2 inhibitor | | Descriptor: | 2-[2-(oxan-4-ylamino)pyrimidin-4-yl]-5-(phenylmethyl)-6,7-dihydro-1~{H}-pyrrolo[3,2-c]pyridin-4-one, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Debreczeni, J.E, Ward, R.A, Bethel, P, Cook, C, Davies, E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, Hudson, J, James, M, Jones, C.D, Jones, C.R, Lamont, S, Lewis, R, Lindsay, N, Roberts, K, Simpson, I, StGallay, S, Swallow, S, Tonge, M. | | Deposit date: | 2017-03-21 | | Release date: | 2017-04-19 | | Last modified: | 2017-05-10 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective Inhibitors of ERK1/2 from a Modestly Active and Promiscuous Chemical Start Point.
J. Med. Chem., 60, 2017
|
|
8AKI
 
 | | Acyl-enzyme complex of ampicillin bound to deacylation mutant KPC-2 (E166Q) | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL, ... | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2022-07-29 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Tautomer-Specific Deacylation and Omega-Loop Flexibility Explain the Carbapenem-Hydrolyzing Broad-Spectrum Activity of the KPC-2 beta-Lactamase.
J.Am.Chem.Soc., 145, 2023
|
|
6GTG
 
 | | Transition state structure of Cpf1(Cas12a) I4 conformation | | Descriptor: | CRISPR-associated endonuclease Cas12a, DNA (32-MER), DNA (5'-D(P*CP*GP*AP*GP*CP*TP*CP*GP*TP*TP*AP*GP*AP*GP*AP*AP*GP*T)-3'), ... | | Authors: | Mesa, P, Montoya, G, Stella, S. | | Deposit date: | 2018-06-18 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Conformational Activation Promotes CRISPR-Cas12a Catalysis and Resetting of the Endonuclease Activity.
Cell, 175, 2018
|
|
5NHJ
 
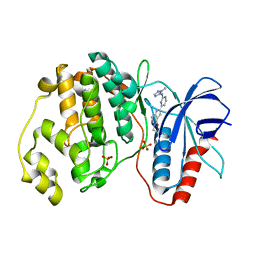 | | Human Erk2 with an Erk1/2 inhibitor | | Descriptor: | 5-(2-methoxyethyl)-2-[2-[(2-methylpyrazol-3-yl)amino]pyrimidin-4-yl]-6,7-dihydro-1~{H}-pyrrolo[3,2-c]pyridin-4-one, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Debreczeni, J.E, Ward, R.A, Bethel, P, Cook, C, Davies, E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, Hudson, J, James, M, Jones, C.D, Jones, C.R, Lamont, S, Lewis, R, Lindsay, N, Roberts, K, Simpson, I, StGallay, S, Swallow, S, Tonge, M. | | Deposit date: | 2017-03-21 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective Inhibitors of ERK1/2 from a Modestly Active and Promiscuous Chemical Start Point.
J. Med. Chem., 60, 2017
|
|
5K6G
 
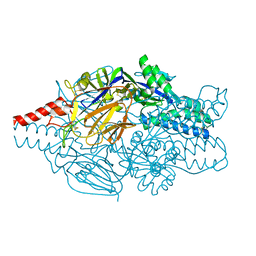 | | Crystal structure of prefusion-stabilized RSV F single-chain 9-24 DS-Cav1 variant. | | Descriptor: | Fusion glycoprotein F0,Fusion glycoprotein F0 | | Authors: | Joyce, M.G, Zhang, B, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2016-05-24 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Iterative structure-based improvement of a fusion-glycoprotein vaccine against RSV.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5NHP
 
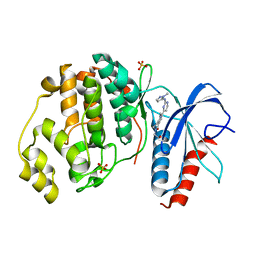 | | Human Erk2 with an Erk1/2 inhibitor | | Descriptor: | 5-(2-methoxyethyl)-1-methyl-2-[2-[(2-methylpyrazol-3-yl)amino]pyrimidin-4-yl]-6,7-dihydropyrrolo[3,2-c]pyridin-4-one, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Debreczeni, J.E, Ward, R.A, Bethel, P, Cook, C, Davies, E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, Hudson, J, James, M, Jones, C.D, Jones, C.R, Lamont, S, Lewis, R, Lindsay, N, Roberts, K, Simpson, I, StGallay, S, Swallow, S, Tonge, M. | | Deposit date: | 2017-03-22 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective Inhibitors of ERK1/2 from a Modestly Active and Promiscuous Chemical Start Point.
J. Med. Chem., 60, 2017
|
|
8AGZ
 
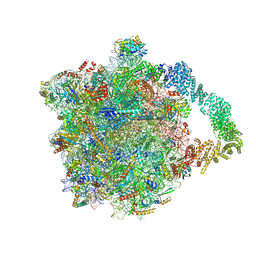 | | Yeast RQC complex in state with the RING domain of Ltn1 in the OUT position | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Tesina, P, Buschauer, R, Beckmann, R. | | Deposit date: | 2022-07-20 | | Release date: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Molecular basis of eIF5A-dependent CAT tailing in eukaryotic ribosome-associated quality control.
Mol.Cell, 83, 2023
|
|
