6JOG
 
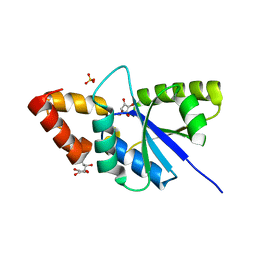 | | Crystal structure of the complex of phospho pantetheine adenylyl transferase from Acinetobacter baumannii with two ascorbic acid (Vitamin-C) molecules. | | Descriptor: | ASCORBIC ACID, Phosphopantetheine adenylyltransferase, SULFATE ION | | Authors: | Viswanathan, V, Gupta, A, Bairagya, H.R, Sharma, S, Singh, T.P. | | Deposit date: | 2019-03-20 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the complex of phospho pantetheine adenylyl transferase from Acinetobacter baumannii with two ascorbic acid (Vitamin-C) molecules.
To Be Published
|
|
4QAJ
 
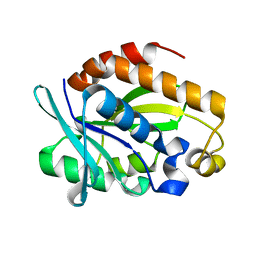 | | Crystal structure of Peptidyl-tRNA hydrolase from Pseudomonas aeruginosa at 1.5 Angstrom resolution | | Descriptor: | Peptidyl-tRNA hydrolase | | Authors: | Singh, A, Kumar, A, Gautam, L, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Arora, A, Singh, T.P. | | Deposit date: | 2014-05-05 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and binding studies of peptidyl-tRNA hydrolase from Pseudomonas aeruginosa provide a platform for the structure-based inhibitor design against peptidyl-tRNA hydrolase
Biochem.J., 463, 2014
|
|
1UMX
 
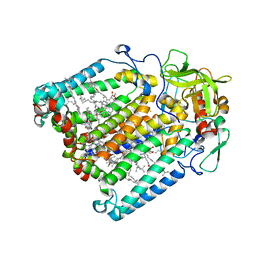 | | PHOTOSYNTHETIC REACTION CENTER MUTANT WITH ARG M267 REPLACED WITH LEU (CHAIN M, R267L) | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN B, FE (III) ION, ... | | Authors: | Fyfe, P.K, Isaacs, N.W, Cogdell, R.J, Jones, M.R. | | Deposit date: | 2003-09-02 | | Release date: | 2004-06-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Disruption of a specific molecular interaction with a bound lipid affects the thermal stability of the purple bacterial reaction centre.
Biochim.Biophys.Acta, 1608, 2004
|
|
4LUM
 
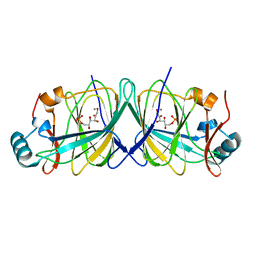 | |
1NF6
 
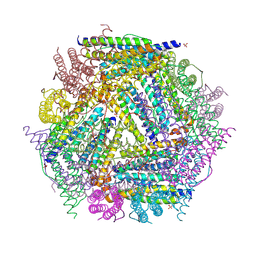 | | X-ray structure of the Desulfovibrio desulfuricans bacterioferritin: the diiron site in different catalytic states ("cycled" structure: reduced in solution and allowed to reoxidise before crystallisation) | | Descriptor: | 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, FE (III) ION, GLYCEROL, ... | | Authors: | Macedo, S, Romao, C.V, Mitchell, E, Matias, P.M, Liu, M.Y, Xavier, A.V, LeGall, J, Teixeira, M, Lindley, P, Carrondo, M.A. | | Deposit date: | 2002-12-13 | | Release date: | 2003-04-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The nature of the di-iron site in the bacterioferritin from
Desulfovibrio desulfuricans
NAT.STRUCT.BIOL., 10, 2003
|
|
7FT1
 
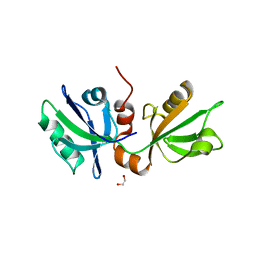 | | SDCBP PanDDA analysis group deposition -- The PDZ domans of SDCBP in complex with Z2204875953 | | Descriptor: | (3P)-3-(2H-1,3-benzodioxol-5-yl)-1H-pyrazole, 1,2-ETHANEDIOL, ALANINE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-01-24 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | SDCBP PanDDA analysis group deposition
To Be Published
|
|
7FT8
 
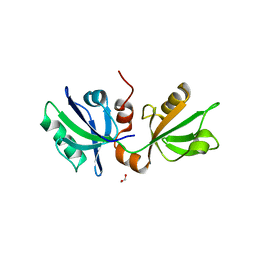 | | SDCBP PanDDA analysis group deposition -- The PDZ domans of SDCBP in complex with NCL-00024671 | | Descriptor: | 1,2-ETHANEDIOL, 2-(4-bromo-1H-pyrazol-1-yl)ethan-1-ol, ALANINE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-01-24 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | SDCBP PanDDA analysis group deposition
To Be Published
|
|
7FSX
 
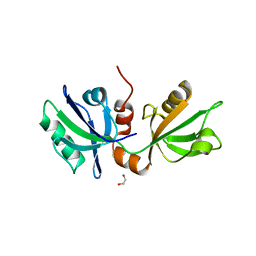 | | SDCBP PanDDA analysis group deposition -- The PDZ domans of SDCBP in complex with Z1328078283 | | Descriptor: | 1,2-ETHANEDIOL, ALANINE, D-GLUTAMIC ACID, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-01-24 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | SDCBP PanDDA analysis group deposition
To Be Published
|
|
1YOW
 
 | | human Steroidogenic Factor 1 LBD with bound Co-factor Peptide | | Descriptor: | PHOSPHATIDYL ETHANOL, Steroidogenic factor 1, TIF2 peptide | | Authors: | Krylova, I.N, Sablin, E.P, Xu, R.X, Waitt, G.M, Juzumiene, D, Williams, J.D, Ingraham, H.A, Willson, T.M, Williams, S.P, Montana, V, Madauss, K.P, Moore, J, Bynum, J.M, Lebedeva, L, MacKay, J.A, Suzawa, M, Guy, R.K, Thornton, J.W. | | Deposit date: | 2005-01-28 | | Release date: | 2005-05-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analyses reveal phosphatidyl inositols as ligands for the NR5 orphan receptors SF-1 and LRH-1
Cell(Cambridge,Mass.), 120, 2005
|
|
7FT5
 
 | | SDCBP PanDDA analysis group deposition -- The PDZ domans of SDCBP in complex with POB0093 | | Descriptor: | 1,2-ETHANEDIOL, 4-(4-methoxyphenyl)oxane-4-carboxylic acid, ALANINE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-01-24 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | SDCBP PanDDA analysis group deposition
To Be Published
|
|
4B0Y
 
 | | Determination of X-ray Structure of human SOUL by Molecular Replacement | | Descriptor: | HEME-BINDING PROTEIN 2 | | Authors: | Freire, F, Carvalho, A.L, Aveiro, S.S, Charbonnier, P, Moulis, J.M, Romao, M.J, Goodfellow, B.J, Macedo, A.L. | | Deposit date: | 2012-07-06 | | Release date: | 2012-08-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Human Soul: A Heme-Binding or a Bh3 Domain-Containing Protein
To be Published
|
|
6J7E
 
 | |
2WYD
 
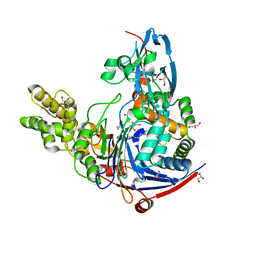 | | The quorum quenching N-acyl homoserine lactone acylase PvdQ in complex with dodecanoic acid | | Descriptor: | ACYL-HOMOSERINE LACTONE ACYLASE PVDQ SUBUNIT ALPHA, ACYL-HOMOSERINE LACTONE ACYLASE PVDQ SUBUNIT BETA, GLYCEROL, ... | | Authors: | Bokhove, M, Nadal Jimenez, P, Quax, W.J, Dijkstra, B.W. | | Deposit date: | 2009-11-16 | | Release date: | 2009-12-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | The Quorum-Quenching N-Acyl Homoserine Lactone Acylase Pvdq is an Ntn-Hydrolase with an Unusual Substrate-Binding Pocket
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1UPD
 
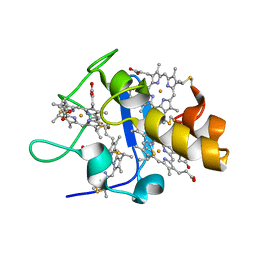 | | Oxidized STRUCTURE OF CYTOCHROME C3 FROM DESULFOVIBRIO DESULFURICANS ATCC 27774 AT PH 7.6 | | Descriptor: | CYTOCHROME C3, HEME C | | Authors: | Bento, I, Matias, P.M, Baptista, A.M, Da Costa, P.N, Van Dongen, W.M.A.M, Saraiva, L.M, Schneider, T.R, Soares, C.M, Carrondo, M.A. | | Deposit date: | 2003-09-29 | | Release date: | 2004-09-30 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular Basis for Redox-Bohr and Cooperative Effects in Cytochrome C3 from Desulfovibrio Desulfuricans Atcc 27774: Crystallographic and Modeling Studies of Oxidized and Reduced High-Resolution Structures at Ph 7.6
Proteins, 54, 2004
|
|
2OTU
 
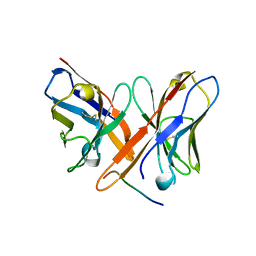 | | Crystal structure of Fv polyglutamine complex | | Descriptor: | Fv heavy chain variable domain, Fv light chain variable domain, peptide antigen | | Authors: | Li, P. | | Deposit date: | 2007-02-09 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Implications of the structure of a poly-Gln/anti-poly-Gln complex for disease progression and therapy
To be Published
|
|
4B2V
 
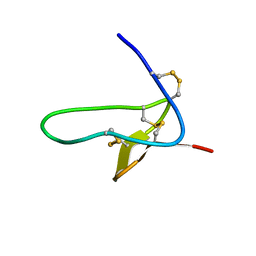 | | S64, a spider venom toxin peptide from Sicarius dolichocephalus | | Descriptor: | S64 | | Authors: | Loening, N.M, Wilson, Z.N, Zobel-Thropp, P.A, Binford, G.J. | | Deposit date: | 2012-07-18 | | Release date: | 2013-01-16 | | Last modified: | 2013-02-06 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of Two Homologous Venom Peptides from Sicarius Dolichocephalus
Plos One, 8, 2013
|
|
6J8V
 
 | | Structure of MOEN5-SSO7D fusion protein in complex with ligand 2 | | Descriptor: | FARNESYL, MoeN5,DNA-binding protein 7d | | Authors: | Ko, T.P, Zhang, L.L, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-01-21 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Complex structures of MoeN5 with substrate analogues suggest sequential catalytic mechanism.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
6AVJ
 
 | | Crystal structure of human Mitochondrial inner NEET protein (MiNT)/CISD3 | | Descriptor: | CDGSH iron-sulfur domain-containing protein 3, mitochondrial, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Lipper, C.H, Karmi, O, Sohn, Y.S, Darash-Yahana, M, Lammert, H, Song, L, Liu, A, Mittler, R, Nechushtai, R, Onuchic, J.N, Jennings, P.A. | | Deposit date: | 2017-09-02 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the human monomeric NEET protein MiNT and its role in regulating iron and reactive oxygen species in cancer cells.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4ARH
 
 | | X ray structure of the periplasmic zinc binding protein ZinT from Salmonella enterica | | Descriptor: | METAL-BINDING PROTEIN YODA, SULFATE ION | | Authors: | Alaleona, F, Ilari, A, Battistoni, A, Petrarca, P, Chiancone, E. | | Deposit date: | 2012-04-24 | | Release date: | 2013-05-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Salmonella Enterica Zint Structure, Zinc Affinity and Interaction with the High-Affinity Uptake Protein Znua Provide Insight Into the Management of Periplasmic Zinc.
Biochim.Biophys.Acta, 1840, 2014
|
|
3GE2
 
 | | Crystal structure of putative lipoprotein SP_0198 from Streptococcus pneumoniae | | Descriptor: | CHLORIDE ION, GLYCEROL, Lipoprotein, ... | | Authors: | Kim, Y, Zhang, R, Joachimiak, G, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-02-24 | | Release date: | 2009-03-17 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Crystal Structure of Putative Lipoprotein SP_0198 from Streptococcus pneumoniae
To be Published
|
|
1C8B
 
 | | CRYSTAL STRUCTURE OF A NOVEL GERMINATION PROTEASE FROM SPORES OF BACILLUS MEGATERIUM: STRUCTURAL REARRANGEMENTS AND ZYMOGEN ACTIVATION | | Descriptor: | SPORE PROTEASE | | Authors: | Ponnuraj, K, Rowland, S, Nessi, C, Setlow, P, Jedrzejas, M.J. | | Deposit date: | 2000-05-03 | | Release date: | 2001-05-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of a novel germination protease from spores of Bacillus megaterium: structural arrangement and zymogen activation.
J.Mol.Biol., 300, 2000
|
|
2WYB
 
 | | The quorum quenching N-acyl homoserine lactone acylase PvdQ with a covalently bound dodecanoic acid | | Descriptor: | ACYL-HOMOSERINE LACTONE ACYLASE PVDQ SUBUNIT ALPHA, ACYL-HOMOSERINE LACTONE ACYLASE PVDQ SUBUNIT BETA, GLYCEROL, ... | | Authors: | Bokhove, M, Nadal Jimenez, P, Quax, W.J, Dijkstra, B.W. | | Deposit date: | 2009-11-16 | | Release date: | 2009-12-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Quorum-Quenching N-Acyl Homoserine Lactone Acylase Pvdq is an Ntn-Hydrolase with an Unusual Substrate-Binding Pocket
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3HU8
 
 | | 2-ethoxyphenol in complex with T4 lysozyme L99A/M102Q | | Descriptor: | 2-ethoxyphenol, Lysozyme, PHOSPHATE ION | | Authors: | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | Deposit date: | 2009-06-13 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
1C8T
 
 | | HUMAN STROMELYSIN-1 (E202Q) CATALYTIC DOMAIN COMPLEXED WITH RO-26-2812 | | Descriptor: | 2-(2-{2-[(BIPHENYL-4-YLMETHYL)-AMINO]-3-MERCAPTO-PENTANOYLAMINO}-ACETYLAMINO)-3-METHYL-BUTYRIC ACID METHYL ESTER, CALCIUM ION, STROMELYSIN-1, ... | | Authors: | Steele, D.L, el-Kabbani, O, Dunten, P, Crowther, R.L. | | Deposit date: | 1999-07-29 | | Release date: | 2000-07-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Expression, characterization and structure determination of an active site mutant (Glu202-Gln) of mini-stromelysin-1.
Protein Eng., 13, 2000
|
|
6B0L
 
 | | KLP10A-AMPPNP in complex with a microtubule | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein Klp10A, ... | | Authors: | Benoit, M.P.M.H, Asenjo, A.B, Sosa, H. | | Deposit date: | 2017-09-14 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Cryo-EM reveals the structural basis of microtubule depolymerization by kinesin-13s.
Nat Commun, 9, 2018
|
|
