1OK6
 
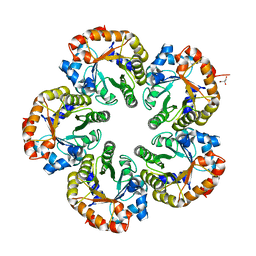 | | Orthorhombic crystal form of an Archaeal fructose 1,6-bisphosphate aldolase | | Descriptor: | FRUCTOSE-BISPHOSPHATE ALDOLASE CLASS I, GLYCEROL | | Authors: | Lorentzen, E, Zwart, P, Stark, A, Hensel, R, Siebers, B, Pohl, E. | | Deposit date: | 2003-07-18 | | Release date: | 2003-09-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of an archaeal class I aldolase and the evolution of (betaalpha)8 barrel proteins.
J. Biol. Chem., 278, 2003
|
|
3W4U
 
 | | Human zeta-2 beta-2-s hemoglobin | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit beta, Hemoglobin subunit zeta, ... | | Authors: | Safo, M.K, Ko, T.-P, Russell, J.E. | | Deposit date: | 2013-01-16 | | Release date: | 2013-02-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of fully liganded Hb zeta 2 beta 2(s) trapped in a tense conformation
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1BCM
 
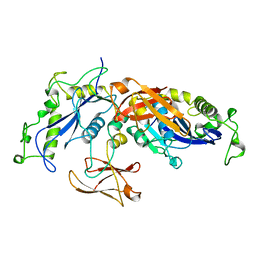 | |
1XLZ
 
 | | Catalytic Domain Of Human Phosphodiesterase 4B In Complex With Filaminast | | Descriptor: | (1E)-1-[3-(CYCLOPENTYLOXY)-4-METHOXYPHENYL]ETHANONE O-(AMINOCARBONYL)OXIME, MAGNESIUM ION, ZINC ION, ... | | Authors: | Card, G.L, England, B.P, Suzuki, Y, Fong, D, Powell, B, Lee, B, Luu, C, Tabrizizad, M, Gillette, S, Ibrahim, P.N, Artis, D.R, Bollag, G, Milburn, M.V, Kim, S.-H, Schlessinger, J, Zhang, K.Y.J. | | Deposit date: | 2004-09-30 | | Release date: | 2004-12-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural Basis for the Activity of Drugs that Inhibit Phosphodiesterases.
STRUCTURE, 12, 2004
|
|
1XJR
 
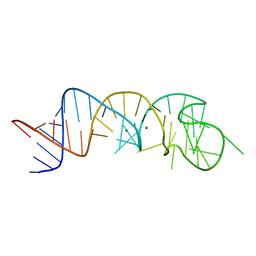 | | The Structure of a Rigorously Conserved RNA Element Within the SARS Virus Genome | | Descriptor: | MAGNESIUM ION, s2m RNA | | Authors: | Robertson, M.P, Igel, H, Baertsch, R, Haussler, D, Ares Jr, M, Scott, W.G. | | Deposit date: | 2004-09-24 | | Release date: | 2005-02-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of a rigorously conserved RNA element within the SARS virus genome
Plos Biol., 3, 2005
|
|
2M62
 
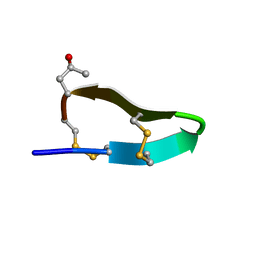 | |
3R1L
 
 | | Crystal structure of the Class I ligase ribozyme-substrate preligation complex, C47U mutant, Mg2+ bound | | Descriptor: | 5'-R(*UP*CP*CP*AP*GP*UP*A)-3', Class I ligase ribozyme, MAGNESIUM ION, ... | | Authors: | Shechner, D.M, Bartel, D.P. | | Deposit date: | 2011-03-10 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.125 Å) | | Cite: | The structural basis of RNA-catalyzed RNA polymerization.
Nat.Struct.Mol.Biol., 18, 2011
|
|
2XVT
 
 | | Structure of the extracellular domain of human RAMP2 | | Descriptor: | CALCIUM ION, RECEPTOR ACTIVITY-MODIFYING PROTEIN 2 | | Authors: | Quigley, A, Pike, A.C.W, Burgess-Brown, N, Krojer, T, Shrestha, L, Goubin, S, Kim, J, Das, S, Muniz, J.R.C, Canning, P, Chaikuad, A, Vollmar, M, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Bountra, C, Barr, A.J, Carpenter, E.P. | | Deposit date: | 2010-10-31 | | Release date: | 2010-12-29 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the Extracellular Domain of Human Ramp2
To be Published
|
|
1U3K
 
 | |
2UVL
 
 | | Human BIR3 domain of Baculoviral Inhibitor of Apoptosis Repeat- Containing 3 (BIRC3) | | Descriptor: | BACULOVIRAL IAP REPEAT-CONTAINING PROTEIN 3, GLYCEROL, ZINC ION | | Authors: | Moche, M, Lehtio, L, Arrowsmith, C, Berglund, H, Busam, R, Collins, R, Dahlgren, L.G, Edwards, A, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg-Schiavone, L, Johansson, I, Karlberg, T, Kotenyova, T, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, Upsten, M, Van Den Berg, S, Weigelt, J, Nordlund, P. | | Deposit date: | 2007-03-12 | | Release date: | 2007-03-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structures of Bir Domains from Human Naip and Ciap2.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
5TEY
 
 | | Human METTL3-METTL14 complex | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, MAGNESIUM ION, ... | | Authors: | DONG, A, ZENG, H, LI, Y, TEMPEL, W, Seitova, A, Hutchinson, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-23 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Human METTL3-METTL14 complex
to be published
|
|
1AOQ
 
 | |
1AOF
 
 | | CYTOCHROME CD1 NITRITE REDUCTASE, REDUCED FORM | | Descriptor: | HEME D, NITRITE REDUCTASE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Williams, P.A, Fulop, V. | | Deposit date: | 1997-07-02 | | Release date: | 1997-10-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Haem-ligand switching during catalysis in crystals of a nitrogen-cycle enzyme.
Nature, 389, 1997
|
|
1OKK
 
 | |
2UW4
 
 | | Structure of PKA-PKB chimera complexed with 2-(4-(5-methyl-1H-pyrazol- 4-yl)-phenyl)-ethylamine | | Descriptor: | 2-[4-(3-METHYL-1H-PYRAZOL-4-YL)PHENYL]ETHANAMINE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA, CAMP-DEPENDENT PROTEIN KINASE, ... | | Authors: | Davies, T.G, Saxty, G, Woodhead, S.J, Berdini, V, Verdonk, M.L, Wyatt, P.G, Boyle, R.G, Barford, D, Downham, R, Garrett, M.D, Carr, R.A. | | Deposit date: | 2007-03-19 | | Release date: | 2007-05-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of Inhibitors of Protein Kinase B Using Fragment-Based Lead Discovery
J.Med.Chem., 50, 2007
|
|
1XQY
 
 | | Crystal structure of F1-mutant S105A complex with PRO-LEU-GLY-GLY | | Descriptor: | PLGG, PROLINE, Proline iminopeptidase | | Authors: | Goettig, P, Brandstetter, H, Groll, M, Goehring, W, Konarev, P.V, Svergun, D.I, Huber, R, Kim, J.-S. | | Deposit date: | 2004-10-13 | | Release date: | 2005-07-12 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-ray snapshots of peptide processing in mutants of tricorn-interacting factor F1 from Thermoplasma acidophilum
J.Biol.Chem., 280, 2005
|
|
2XUG
 
 | | Crystal structure of mAChE-Y337A-TZ2PA6 anti complex (1 wk) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,8-DIAMINO-6-PHENYL-5-[6-[1-[2-[(1,2,3,4-TETRAHYDRO-9-ACRIDINYL)AMINO]ETHYL]-1H-1,2,3-TRIAZOL-4-YL]HEXYL]-PHENANTHRIDINIUM, ACETYLCHOLINESTERASE, ... | | Authors: | Bourne, Y, Radic, Z, Taylor, P, Marchot, P. | | Deposit date: | 2010-10-19 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conformational Remodeling of Femtomolar Inhibitor-Acetylcholinesterase Complexes in the Crystalline State
J.Am.Chem.Soc., 132, 2010
|
|
1GY9
 
 | | Taurine/alpha-ketoglutarate Dioxygenase from Escherichia coli | | Descriptor: | 2-AMINOETHANESULFONIC ACID, 2-OXOGLUTARIC ACID, ALPHA-KETOGLUTARATE-DEPENDENT TAURINE DIOXYGENASE, ... | | Authors: | Elkins, J.M, Burzlaff, N.I, Ryle, M.J, Lloyd, J.S, Clifton, I.J, Baldwin, J.E, Hausinger, R.P, Roach, P.L. | | Deposit date: | 2002-04-22 | | Release date: | 2002-04-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-Ray Crystal Structure of Escherichia Coli Taurine/Alpha-Ketoglutarate Dioxygenase Complexed to Ferrous Iron and Substrates.
Biochemistry, 41, 2002
|
|
1BFZ
 
 | | BOUND CONFORMATION OF N-TERMINAL CLEAVAGE PRODUCT PEPTIDE MIMIC (P1-P9 OF RELEASE SITE) WHILE BOUND TO HCMV PROTEASE AS DETERMINED BY TRANSFERRED NOESY EXPERIMENTS (P1-P5 SHOWN ONLY), NMR, 32 STRUCTURES | | Descriptor: | HCMV PROTEASE R-SITE N-TERMINAL CLEAVAGE PRODUCT | | Authors: | Laplante, S.R, Aubry, N, Bonneau, P.R, Cameron, D.R, Lagace, L, Massariol, M.-J, Montpetit, H, Ploufe, C, Kawai, S.H, Fulton, B.D, Chen, Z, Ni, F. | | Deposit date: | 1998-05-25 | | Release date: | 1999-05-25 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Human cytomegalovirus protease complexes its substrate recognition sequences in an extended peptide conformation.
Biochemistry, 37, 1998
|
|
2UW5
 
 | | Structure of PKA-PKB chimera complexed with (R)-2-(4-chloro-phenyl)- 2-(4-1H-pyrazol-4-yl)-phenyl)-ethylamine | | Descriptor: | (2R)-2-(4-CHLOROPHENYL)-2-[4-(1H-PYRAZOL-4-YL)PHENYL]ETHANAMINE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA, CAMP-DEPENDENT PROTEIN KINASE, ... | | Authors: | Davies, T.G, Saxty, G, Woodhead, S.J, Berdini, V, Verdonk, M.L, Wyatt, P.G, Boyle, R.G, Barford, D, Downham, R, Garrett, M.D, Carr, R.A. | | Deposit date: | 2007-03-19 | | Release date: | 2007-05-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Identification of Inhibitors of Protein Kinase B Using Fragment-Based Lead Discovery
J.Med.Chem., 50, 2007
|
|
1AOM
 
 | |
1E0F
 
 | | Crystal structure of the human alpha-thrombin-haemadin complex: an exosite II-binding inhibitor | | Descriptor: | HAEMADIN, THROMBIN | | Authors: | Richardson, J.L, Kroeger, B, Hoefken, W, Pereira, P, Huber, R, Bode, W, Fuentes-Prior, P. | | Deposit date: | 2000-03-27 | | Release date: | 2000-11-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of the Human Alpha-Thrombin-Haemadin Complex: An Exosite II-Binding Inhibitor
Embo J., 19, 2000
|
|
2UW7
 
 | | Structure of PKA-PKB chimera complexed with 4-(4-chloro-phenyl)-4-(4- (1H-pyrazol-4-yl)-phenyl)-piperidine | | Descriptor: | 4-(4-CHLOROPHENYL)-4-[4-(1H-PYRAZOL-4-YL)PHENYL]PIPERIDINE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA, CAMP-DEPENDENT PROTEIN KINASE, ... | | Authors: | Davies, T.G, Saxty, G, Woodhead, S.J, Berdini, V, Verdonk, M.L, Wyatt, P.G, Boyle, R.G, Barford, D, Downham, R, Garrett, M.D, Carr, R.A. | | Deposit date: | 2007-03-19 | | Release date: | 2007-05-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of Inhibitors of Protein Kinase B Using Fragment-Based Lead Discovery
J.Med.Chem., 50, 2007
|
|
1XOT
 
 | | Catalytic Domain Of Human Phosphodiesterase 4B In Complex With Vardenafil | | Descriptor: | 2-{2-ETHOXY-5-[(4-ETHYLPIPERAZIN-1-YL)SULFONYL]PHENYL}-5-METHYL-7-PROPYLIMIDAZO[5,1-F][1,2,4]TRIAZIN-4(1H)-ONE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Card, G.L, England, B.P, Suzuki, Y, Fong, D, Powell, B, Lee, B, Luu, C, Tabrizizad, M, Gillette, S, Ibrahim, P.N, Artis, D.R, Bollag, G, Milburn, M.V, Kim, S.-H, Schlessinger, J, Zhang, K.Y.J. | | Deposit date: | 2004-10-06 | | Release date: | 2004-12-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural Basis for the Activity of Drugs that Inhibit Phosphodiesterases.
STRUCTURE, 12, 2004
|
|
1XQX
 
 | | Crystal structure of F1-mutant S105A complex with PCK | | Descriptor: | PHENYLALANYLMETHYLCHLORIDE, Proline iminopeptidase | | Authors: | Goettig, P, Brandstetter, H, Groll, M, Goehring, W, Konarev, P.V, Svergun, D.I, Huber, R, Kim, J.-S. | | Deposit date: | 2004-10-13 | | Release date: | 2005-07-12 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray snapshots of peptide processing in mutants of tricorn-interacting factor F1 from Thermoplasma acidophilum
J.Biol.Chem., 280, 2005
|
|
