8HQE
 
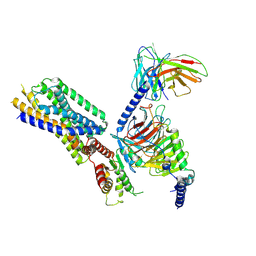 | | Cryo-EM structure of the apo-GPR132-Gi | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wang, J.L, Ding, J.H, Sun, J.P, Yu, X. | | Deposit date: | 2022-12-13 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Functional screening and rational design of compounds targeting GPR132 to treat diabetes.
Nat Metab, 5, 2023
|
|
3PO6
 
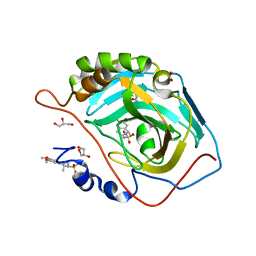 | | Crystal structure of human carbonic anhydrase II with 6,7-Dimethoxy-1-methyl-3,4-dihydroisoquinoline-2(1H)-sulfonamide | | Descriptor: | (1R)-6,7-dimethoxy-1-methyl-3,4-dihydroisoquinoline-2(1H)-sulfonamide, (1S)-6,7-dimethoxy-1-methyl-3,4-dihydroisoquinoline-2(1H)-sulfonamide, ACETATE ION, ... | | Authors: | Mader, P, Brynda, J, Gitto, R, Agnello, S, Ferro, S, De Luca, L, Vullo, D, Supuran, C.T, Chimirri, A. | | Deposit date: | 2010-11-22 | | Release date: | 2011-04-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural basis for the interaction between carbonic anhydrase and 1,2,3,4-tetrahydroisoquinolin-2-ylsulfonamides.
J.Med.Chem., 54, 2011
|
|
4WOG
 
 | | Crystal Structure of Frutalin from Artocarpus incisa | | Descriptor: | Frutalin | | Authors: | Pereira, H.M, Moreira, A.C.O.M, Vieira Neto, A.E, Moreno, F.B.M.B, Lobo, M.D.P, Sousa, F.D, Grangeiro, T.B, Moreira, R.A. | | Deposit date: | 2014-10-15 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.813 Å) | | Cite: | Crystal Structure of Frutalin from Artocarpus incisa
To Be Published
|
|
4ZJ3
 
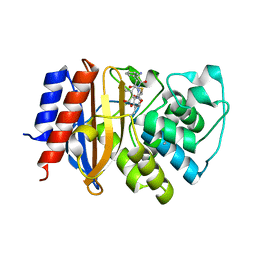 | | Crystal structure of cephalexin bound acyl-enzyme intermediate of Val216AcrF mutant TEM1 beta-lactamase from Escherichia coli: E166N and V216AcrF mutant. | | Descriptor: | Beta-lactamase TEM | | Authors: | Xiao, H, Nasertorabi, F, Choi, S, Han, G.W, Reed, S.A, Stevens, R.C, Schultz, P.G. | | Deposit date: | 2015-04-29 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Exploring the potential impact of an expanded genetic code on protein function.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3EIK
 
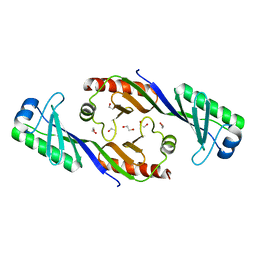 | |
4WF5
 
 | | Crystal structure of E.Coli DsbA soaked with compound 4 | | Descriptor: | 1,2-ETHANEDIOL, 4-methyl-2-[4-(trifluoromethyl)phenyl]-1,3-thiazole-5-carboxylic acid, COPPER (II) ION, ... | | Authors: | Adams, L.A, Sharma, P, Mohanty, B, Ilyichova, O.V, Mulcair, M.D, Williams, M.L, Gleeson, E.C, Totsika, M, Doak, B.C, Caria, S, Rimmer, K, Shouldice, S.R, Vazirani, M, Headey, S.J, Plumb, B.R, Martin, J.L, Heras, B, Simpson, J.S, Scanlon, M.J. | | Deposit date: | 2014-09-12 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Application of Fragment-Based Screening to the Design of Inhibitors of Escherichia coli DsbA.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
8HQM
 
 | | Activation mechanism of GPR132 by NPGLY | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wang, J.L, Ding, J.H, Sun, J.P, Yu, X. | | Deposit date: | 2022-12-13 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Functional screening and rational design of compounds targeting GPR132 to treat diabetes.
Nat Metab, 5, 2023
|
|
3EJ3
 
 | | Structural and mechanistic analysis of trans-3-chloroacrylic acid dehalogenase activity | | Descriptor: | ACETATE ION, Alpha-subunit of trans-3-chloroacrylic acid dehalogenase, Beta-subunit of trans-3-chloroacrylic acid dehalogenase, ... | | Authors: | Pegan, S, Serrano, H, Whitman, C.P, Mesecar, A.D. | | Deposit date: | 2008-09-17 | | Release date: | 2008-12-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and mechanistic analysis of trans-3-chloroacrylic acid dehalogenase activity.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
6XSD
 
 | | Patient-derived B2GPI | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-glycoprotein 1 | | Authors: | Klenotic, P.A, Yu, E.W.Y. | | Deposit date: | 2020-07-15 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | B2-Glycoprotein I and it's role in APS
To Be Published
|
|
1AGT
 
 | | SOLUTION STRUCTURE OF THE POTASSIUM CHANNEL INHIBITOR AGITOXIN 2: CALIPER FOR PROBING CHANNEL GEOMETRY | | Descriptor: | AGITOXIN 2 | | Authors: | Krezel, A.M, Kasibhatla, C, Hidalgo, P, Mackinnon, R, Wagner, G. | | Deposit date: | 1995-04-14 | | Release date: | 1995-07-10 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the potassium channel inhibitor agitoxin 2: caliper for probing channel geometry.
Protein Sci., 4, 1995
|
|
8HBG
 
 | | FMDV (A/TUR/14/98) in complex with M678F | | Descriptor: | M678F nab, VP1 of capsid protein, VP2 of capsid protein, ... | | Authors: | Li, H.Z, Dong, H, Liu, P. | | Deposit date: | 2022-10-28 | | Release date: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural and in vivo studies of neutralizing antibody topographical classifications reveal mechanisms underlying differences in immunogenicity and antigenicity between 146S and 12S of foot-and-mouth disease virus
To Be Published
|
|
3PQ5
 
 | | Structure of I274C variant of E. coli KatE[] - Images 19-24 | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE 17R, 18S, ... | | Authors: | Loewen, P.C, Jha, V, Louis, S, Chelikani, P, Carpena, X, Fita, I. | | Deposit date: | 2010-11-25 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modulation of heme orientation and binding by a single residue in catalase HPII of Escherichia coli.
Biochemistry, 50, 2011
|
|
1WZ9
 
 | | The 2.1 A structure of a tumour suppressing serpin | | Descriptor: | Maspin precursor, SULFATE ION | | Authors: | Law, R.H, Irving, J.A, Buckle, A.M, Ruzyla, K, Buzza, M, Bashtannyk-Puhalovich, T.A, Beddoe, T.C, Kim, N, Worrall, D.M, Bottomley, S.P, Bird, P.I, Rossjohn, J, Whisstock, J.C. | | Deposit date: | 2005-03-03 | | Release date: | 2005-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The high resolution crystal structure of the human tumor suppressor maspin reveals a novel conformational switch in the G-helix.
J.Biol.Chem., 280, 2005
|
|
6OMC
 
 | | capsid of T5 virion | | Descriptor: | Major capsid protein | | Authors: | Huet, A, Duda, R.L, Boulanger, P, Conway, J.F. | | Deposit date: | 2019-04-18 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Capsid expansion of bacteriophage T5 revealed by high resolution cryoelectron microscopy.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
2LZW
 
 | | DNA duplex containing mispair-aligned O6G-heptylene-O6G interstrand cross-link | | Descriptor: | DNA (5'-D(*CP*GP*AP*AP*AP*GP*TP*TP*TP*CP*G)-3'), HEPTANE | | Authors: | Denisov, A.Y, McManus, F.P, Noronha, A.M, Wilds, C.J. | | Deposit date: | 2012-10-11 | | Release date: | 2013-10-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of interstrand cross-link repair by O 6 -alkylguanine DNA alkyltransferase.
Org.Biomol.Chem., 15, 2017
|
|
2BGU
 
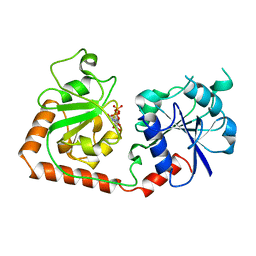 | | CRYSTAL STRUCTURE OF THE DNA MODIFYING ENZYME BETA-GLUCOSYLTRANSFERASE IN THE PRESENCE AND ABSENCE OF THE SUBSTRATE URIDINE DIPHOSPHOGLUCOSE | | Descriptor: | BETA-GLUCOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE | | Authors: | Vrielink, A, Rueger, W, Driessen, H.P.C, Freemont, P.S. | | Deposit date: | 1994-06-09 | | Release date: | 1995-12-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the DNA modifying enzyme beta-glucosyltransferase in the presence and absence of the substrate uridine diphosphoglucose.
EMBO J., 13, 1994
|
|
6XST
 
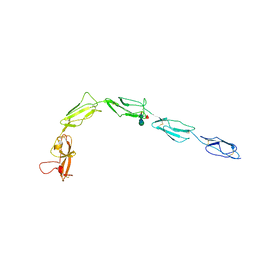 | |
1A9B
 
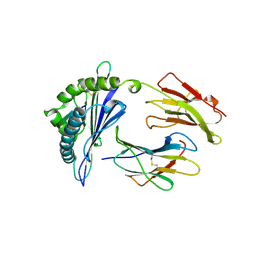 | | DECAMER-LIKE CONFORMATION OF A NANO-PEPTIDE BOUND TO HLA-B3501 DUE TO NONSTANDARD POSITIONING OF THE C-TERMINUS | | Descriptor: | BETA-2-MICROGLOBULIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, B-35 B*3501 (ALPHA CHAIN), ... | | Authors: | Menssen, R, Orth, P, Ziegler, A, Saenger, W. | | Deposit date: | 1998-04-03 | | Release date: | 1998-10-21 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Decamer-like conformation of a nona-peptide bound to HLA-B*3501 due to non-standard positioning of the C terminus.
J.Mol.Biol., 285, 1999
|
|
2BJJ
 
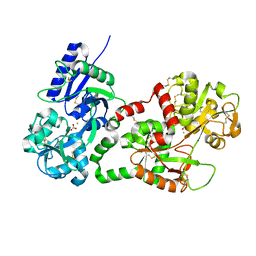 | | Structure of recombinant human lactoferrin produced in the milk of transgenic cows | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, FE (III) ION, ... | | Authors: | Thomassen, E.A.J, Van Veen, H.A, Van Berkel, P.H.C, Nuijens, J.H, Abrahams, J.P. | | Deposit date: | 2005-02-03 | | Release date: | 2005-08-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Protein Structure of Recombinant Human Lactoferrin Produced in the Milk of Transgenic Cows Closely Matches the Structure of Human Milk-Derived Lactoferrin
Transgenic Res., 14, 2005
|
|
1AN0
 
 | | CDC42HS-GDP COMPLEX | | Descriptor: | CDC42HS-GDP, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Kongsaeree, P, Cerione, R, Clardy, J. | | Deposit date: | 1997-06-26 | | Release date: | 1999-01-13 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Structure Determination of Cdc42Hs and Gdp Complex
To be Published
|
|
3PUN
 
 | | Crystal structure of P domain dimer of Norovirus VA207 with Lewis y tetrasaccharide | | Descriptor: | Capsid, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Chen, Y, Tan, M, Xia, M, Hao, N, Zhang, X.C, Huang, P, Jiang, X, Li, X, Rao, Z. | | Deposit date: | 2010-12-06 | | Release date: | 2011-08-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystallography of a Lewis-binding norovirus, elucidation of strain-specificity to the polymorphic human histo-blood group antigens
Plos Pathog., 7, 2011
|
|
2C44
 
 | | Crystal Structure of E. coli Tryptophanase | | Descriptor: | POTASSIUM ION, SULFATE ION, TRYPTOPHANASE | | Authors: | Ku, S.-Y, Yip, P, Howell, P.L. | | Deposit date: | 2005-10-15 | | Release date: | 2006-06-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Escherichia Coli Tryptophanase
Acta Crystallogr.,Sect.D, 62, 2006
|
|
5KJD
 
 | |
4ZQB
 
 | | Crystal structure of NADP-dependent dehydrogenase from Rhodobactersphaeroides in complex with NADP and sulfate | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Kowiel, M, Gasiorowska, O.A, Shabalin, I.G, Handing, K.B, Porebski, P.J, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-05-08 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of NADP-dependent dehydrogenase from Rhodobactersphaeroides in complex with NADP and sulfate
to be published
|
|
5KK0
 
 | | Synechocystis ACO mutant - T136A | | Descriptor: | Apocarotenoid-15,15'-oxygenase, CHLORIDE ION, FE (II) ION | | Authors: | Sui, X, Kiser, P.D, Palczewski, K. | | Deposit date: | 2016-06-20 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Utilization of Dioxygen by Carotenoid Cleavage Oxygenases.
J.Biol.Chem., 290, 2015
|
|
