3ORS
 
 | | Crystal Structure of N5-Carboxyaminoimidazole Ribonucleotide Mutase from Staphylococcus aureus | | Descriptor: | N5-Carboxyaminoimidazole Ribonucleotide Mutase, SULFATE ION | | Authors: | Brugarolas, P, Duguid, E.M, Zhang, W, Poor, C.B, He, C. | | Deposit date: | 2010-09-07 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and biochemical characterization of N5-carboxyaminoimidazole ribonucleotide synthetase and N5-carboxyaminoimidazole ribonucleotide mutase from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
1ADV
 
 | | EARLY E2A DNA-BINDING PROTEIN | | Descriptor: | ADENOVIRUS SINGLE-STRANDED DNA-BINDING PROTEIN, ZINC ION | | Authors: | Kanellopoulos, P.N, Tsernoglou, D, Van Der Vliet, P.C, Tucker, P.A. | | Deposit date: | 1995-05-12 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Alternative arrangements of the protein chain are possible for the adenovirus single-stranded DNA binding protein.
J.Mol.Biol., 257, 1996
|
|
2V40
 
 | | Human Adenylosuccinate synthetase isozyme 2 in complex with GDP | | Descriptor: | ADENYLOSUCCINATE SYNTHETASE ISOZYME 2, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Welin, M, Moche, M, Arrowsmith, C, Berglund, H, Busam, R, Collins, R, Dahlgren, L.G, Edwards, A, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg-Schiavone, L, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Lehtio, L, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, Tresaugues, L, van den Berg, S, Weigelt, J, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-27 | | Release date: | 2007-07-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Human Adenylosuccinate Synthetase Isozyme 2 in Complex with Gdp
To be Published
|
|
8E63
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a phenyl sulfane inhibitor | | Descriptor: | (1R,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-[(N-{[2-(phenylsulfanyl)ethoxy]carbonyl}-L-leucyl)amino]propane-1-sulfonic acid, 2-phenylsulfanylethyl ~{N}-[(2~{S})-1-[[(1~{S},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate, 3C-like proteinase, ... | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Madden, T.K, Groutas, W.C. | | Deposit date: | 2022-08-22 | | Release date: | 2022-09-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-guided design of direct-acting antivirals that exploit the gem-dimethyl effect and potently inhibit 3CL proteases of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) and middle east respiratory syndrome coronavirus (MERS-CoV).
Eur.J.Med.Chem., 254, 2023
|
|
7UOP
 
 | |
7UP9
 
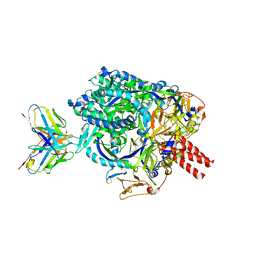 | | Prefusion-stabilized Nipah virus fusion protein complexed with Fab 2D3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 2D3 heavy chain, ... | | Authors: | Byrne, P.O, McLellan, J.S. | | Deposit date: | 2022-04-14 | | Release date: | 2023-03-29 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for antibody recognition of vulnerable epitopes on Nipah virus F protein.
Nat Commun, 14, 2023
|
|
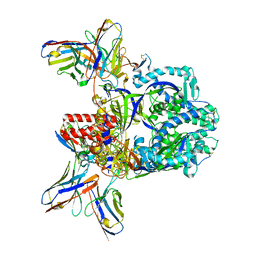
7UPD
 
 | |
1TG1
 
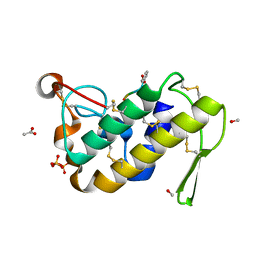 | | Crystal Structure of the complex formed between russells viper phospholipase A2 and a designed peptide inhibitor PHQ-Leu-Val-Arg-Tyr at 1.2A resolution | | Descriptor: | ACETIC ACID, METHANOL, Phospholipase A2, ... | | Authors: | Singh, N, Kaur, P, Somvanshi, R.K, Sharma, S, Dey, S, Perbandt, M, Betzel, C, Singh, T.P. | | Deposit date: | 2004-05-28 | | Release date: | 2004-06-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal Structure of the complex formed between russells viper phospholipase A2 and a designed peptide inhibitor Cbz-dehydro-Leu-Val-Arg-Tyr at 1.2A resolution
To be Published
|
|
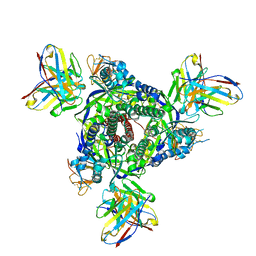
7UPA
 
 | |
3SDF
 
 | | Crystal Structure of C-lobe of Bovine lactoferrin Complexed with Lipoteichoic acid at 2.1 A Resolution | | Descriptor: | (2S)-1-({3-O-[2-(acetylamino)-4-amino-2,4,6-trideoxy-beta-D-galactopyranosyl]-alpha-D-glucopyranosyl}oxy)-3-(heptanoyloxy)propan-2-yl (7Z)-pentadec-7-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shukla, P.K, Gautam, L, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-06-09 | | Release date: | 2011-06-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of C-lobe of Bovine lactoferrin Complexed with Lipoteichoic acid at 2.1 A Resolution
To be Published
|
|
4L7T
 
 | | B. fragilis NanU | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, NanU sialic acid binding protein, ... | | Authors: | Rafferty, J.B, Phansopa, C, Stafford, G.P. | | Deposit date: | 2013-06-14 | | Release date: | 2014-01-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural and functional characterization of NanU, a novel high-affinity sialic acid-inducible binding protein of oral and gut-dwelling Bacteroidetes species.
Biochem.J., 458, 2014
|
|
7FSV
 
 | | SDCBP PanDDA analysis group deposition -- The PDZ domans of SDCBP in complex with Z1454310449 | | Descriptor: | 1,2-ETHANEDIOL, ALANINE, D-GLUTAMIC ACID, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-01-24 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | SDCBP PanDDA analysis group deposition
To Be Published
|
|
5CX9
 
 | | Crystal structure of CK2alpha with (methyl 4-((3-(3-chloro-4-(phenyl)benzylamino)propyl)amino)-4-oxobutanoate bound | | Descriptor: | ACETATE ION, Casein kinase II subunit alpha, PHOSPHATE ION, ... | | Authors: | Brear, P, De Fusco, C, Georgiou, K.H, Spring, D, Hyvonen, M. | | Deposit date: | 2015-07-28 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.732 Å) | | Cite: | A fragment-based approach leading to the discovery of a novel binding site and the selective CK2 inhibitor CAM4066.
Bioorg. Med. Chem., 25, 2017
|
|
3P1D
 
 | | Crystal structure of the bromodomain of human CREBBP in complex with N-Methyl-2-pyrrolidone (NMP) | | Descriptor: | 1-methylpyrrolidin-2-one, CREB-binding protein, POTASSIUM ION, ... | | Authors: | Filippakopoulos, P, Picaud, S, Feletar, I, Fedorov, O, Muniz, J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-09-30 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
7FSR
 
 | | SDCBP PanDDA analysis group deposition -- The PDZ domans of SDCBP in complex with Z54615640 | | Descriptor: | 1,2-ETHANEDIOL, ALANINE, D-GLUTAMIC ACID, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-01-24 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | SDCBP PanDDA analysis group deposition
To Be Published
|
|
1L7H
 
 | | Crystal structure of R292K mutant influenza virus neuraminidase in complex with BCX-1812 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(1-ACETYLAMINO-2-ETHYL-BUTYL)-4-GUANIDINO-2-HYDROXY-CYCLOPENTANECARBOXYLIC ACID, ... | | Authors: | Smith, B.J, McKimm-Breshkin, J.L, McDonald, M, Fernley, R.T, Varghese, J.N, Colman, P.M. | | Deposit date: | 2002-03-15 | | Release date: | 2002-05-29 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural studies of the resistance of influenza virus neuramindase to inhibitors.
J.Med.Chem., 45, 2002
|
|
5CZ2
 
 | | Crystal structure of a two-domain fragment of MMTV integrase | | Descriptor: | MAGNESIUM ION, Pol polyprotein, ZINC ION | | Authors: | Cook, N, Ballandras-Colas, A, Engelman, A, Cherepanov, P. | | Deposit date: | 2015-07-31 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Cryo-EM reveals a novel octameric integrase structure for betaretroviral intasome function.
Nature, 530, 2016
|
|
7FT9
 
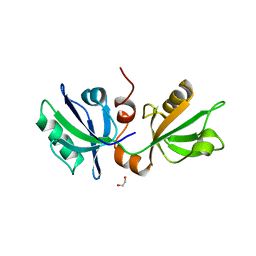 | | SDCBP PanDDA analysis group deposition -- The PDZ domans of SDCBP in complex with Z45636695 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(METHYLSULFONYL)AMINO]BENZOIC ACID, ALANINE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-01-24 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | SDCBP PanDDA analysis group deposition
To Be Published
|
|
7FTA
 
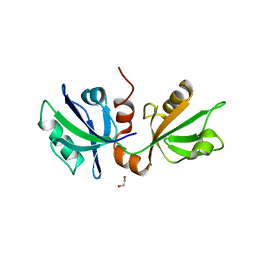 | | SDCBP PanDDA analysis group deposition -- The PDZ domans of SDCBP in complex with Z228589380 | | Descriptor: | 1,2-ETHANEDIOL, 3-[(pyrimidin-2-yl)amino]benzoic acid, D-GLUTAMIC ACID, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-01-24 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | SDCBP PanDDA analysis group deposition
To Be Published
|
|
7FSJ
 
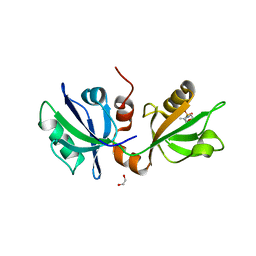 | | SDCBP PanDDA analysis group deposition -- The PDZ domans of SDCBP in complex with Z57744712 | | Descriptor: | 1,2-ETHANEDIOL, ALANINE, D-GLUTAMIC ACID, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-01-24 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | SDCBP PanDDA analysis group deposition
To Be Published
|
|
7FSU
 
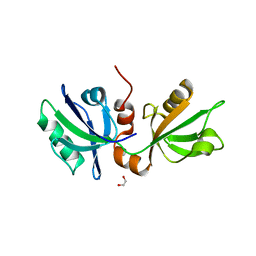 | | SDCBP PanDDA analysis group deposition -- The PDZ domans of SDCBP in complex with Z729726784 | | Descriptor: | 1,2-ETHANEDIOL, 2-cyclopentyl-N-(3-methyl-1,2,4-oxadiazol-5-yl)acetamide, ALANINE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-01-24 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | SDCBP PanDDA analysis group deposition
To Be Published
|
|
7FTB
 
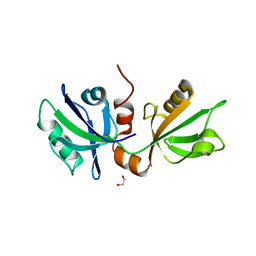 | | SDCBP PanDDA analysis group deposition -- The PDZ domans of SDCBP in complex with Z1198269757 | | Descriptor: | 1,2-ETHANEDIOL, D-GLUTAMIC ACID, GLYCINE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-01-24 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | SDCBP PanDDA analysis group deposition
To Be Published
|
|
7FST
 
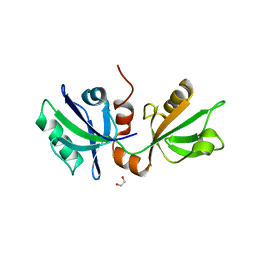 | | SDCBP PanDDA analysis group deposition -- The PDZ domans of SDCBP in complex with Z26968795 | | Descriptor: | 1,2-ETHANEDIOL, ALANINE, D-GLUTAMIC ACID, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-01-24 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | SDCBP PanDDA analysis group deposition
To Be Published
|
|
7FSM
 
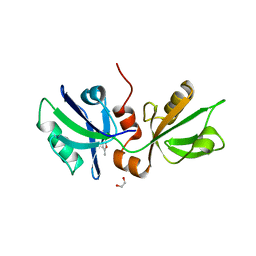 | | SDCBP PanDDA analysis group deposition -- The PDZ domans of SDCBP in complex with Z763030030 | | Descriptor: | 1,2-ETHANEDIOL, ALANINE, D-GLUTAMIC ACID, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-01-24 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | SDCBP PanDDA analysis group deposition
To Be Published
|
|
1TJA
 
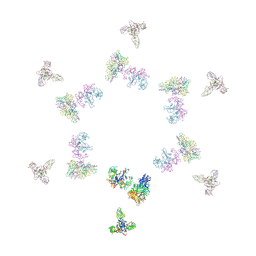 | | Fitting of gp8, gp9, and gp11 into the cryo-EM reconstruction of the bacteriophage T4 contracted tail | | Descriptor: | Baseplate structural protein Gp11, Baseplate structural protein Gp8, Baseplate structural protein Gp9 | | Authors: | Leiman, P.G, Chipman, P.R, Kostyuchenko, V.A, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2004-06-03 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (16 Å) | | Cite: | Three-dimensional rearrangement of proteins in the tail of bacteriophage t4 on infection of its host
Cell(Cambridge,Mass.), 118, 2004
|
|
