1O49
 
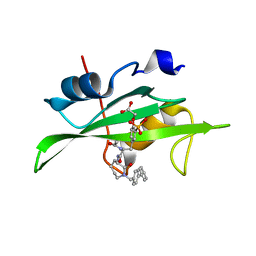 | | CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU85493. | | Descriptor: | PROTO-ONCOGENE TYROSINE-PROTEIN KINASE SRC, {4-[2-ACETYLAMINO-2-(1-BIPHENYL-4-YLMETHYL-2-OXO-AZEPAN-3-YLCARBAMOYL)-ETHYL]-2-PHOSPHONO-PHENOXY}-ACETIC ACID | | Authors: | Lange, G, Loenze, P, Liesum, A. | | Deposit date: | 2003-06-15 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Requirements for specific binding of low affinity inhibitor fragments to the SH2 domain of (pp60)Src are identical to those for high affinity binding of full length inhibitors.
J.Med.Chem., 46, 2003
|
|
7ANM
 
 | | Nudaurelia capensis omega virus capsid: virus-like particles expressed in Nicotiana benthamiana | | Descriptor: | p70 | | Authors: | Castells-Graells, R, Ribeiro, J.R.S, Domitrovic, T, Hesketh, E.L, Scarff, C.A, Johnson, J.E, Ranson, N.A, Lawson, D.M, Lomonossoff, G.P. | | Deposit date: | 2020-10-12 | | Release date: | 2021-08-25 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Plant-expressed virus-like particles reveal the intricate maturation process of a eukaryotic virus.
Commun Biol, 4, 2021
|
|
6EU2
 
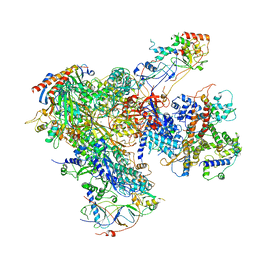 | | Apo RNA Polymerase III - open conformation (oPOL3) | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Abascal-Palacios, G, Ramsay, E.P, Beuron, F, Morris, E, Vannini, A. | | Deposit date: | 2017-10-27 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of RNA polymerase III transcription initiation.
Nature, 553, 2018
|
|
1O4G
 
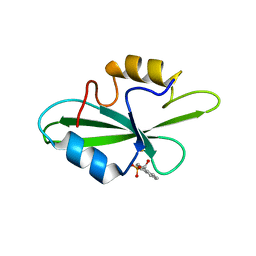 | | CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH DPI59. | | Descriptor: | HYDROXY(1-NAPHTHYL)METHYLPHOSPHONIC ACID, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE SRC | | Authors: | Lange, G, Loenze, P, Liesum, A. | | Deposit date: | 2003-06-15 | | Release date: | 2004-02-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Requirements for specific binding of low affinity inhibitor fragments to the SH2 domain of (pp60)Src are identical to those for high affinity binding of full length inhibitors.
J.Med.Chem., 46, 2003
|
|
6STU
 
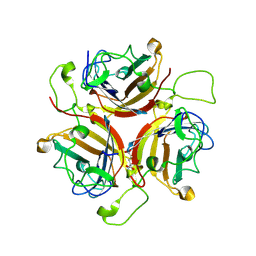 | | Adenovirus 30 Fiber Knob protein | | Descriptor: | 1,2-ETHANEDIOL, Fiber, GLUTAMIC ACID | | Authors: | Baker, A.T, Mundy, R.M, Rizkallah, P.J, Parker, A.L. | | Deposit date: | 2019-09-11 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Broad sialic acid usage amongst species D human adenovirus
npj Viruses, 1, 2023
|
|
7ATA
 
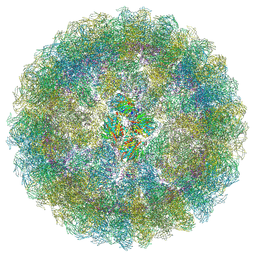 | | Nudaurelia capensis omega virus procapsid: virus-like particles expressed in Nicotiana benthamiana | | Descriptor: | p70 | | Authors: | Castells-Graells, R, Ribeiro, J.R.S, Domitrovic, T, Hesketh, E.L, Scarff, C.A, Johnson, J.E, Ranson, N.A, Lawson, D.M, Lomonossoff, G.P. | | Deposit date: | 2020-10-29 | | Release date: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (6.63 Å) | | Cite: | Plant-expressed virus-like particles reveal the intricate maturation process of a eukaryotic virus.
Commun Biol, 4, 2021
|
|
1O4M
 
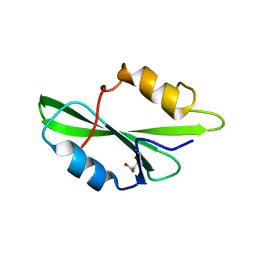 | | CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH MALONICACID. | | Descriptor: | MALONIC ACID, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE SRC | | Authors: | Lange, G, Loenze, P, Liesum, A. | | Deposit date: | 2003-06-15 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Requirements for specific binding of low affinity inhibitor fragments to the SH2 domain of (pp60)Src are identical to those for high affinity binding of full length inhibitors.
J.Med.Chem., 46, 2003
|
|
7AI3
 
 | |
6VGI
 
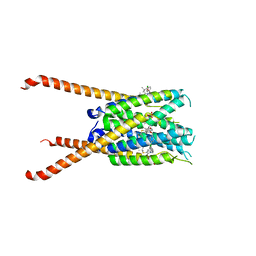 | | Crystal Structures of FLAP bound to MK-866 | | Descriptor: | 3-[3-(tert-butylsulfanyl)-1-[(4-chlorophenyl)methyl]-5-(propan-2-yl)-1H-indol-2-yl]-2,2-dimethylpropanoic acid, 5-lipoxygenase-activating protein, SULFATE ION | | Authors: | Ho, J.D, Lee, M.R, Rauch, C.T, Aznavour, K, Park, J.S, Luz, J.G, Antonysamy, S, Condon, B, Maletic, M, Zhang, A, Hickey, M.J, Hughes, N.E, Chandrasekhar, S, Sloan, A.V, Gooding, K, Harvey, A, Yu, X.P, Kahl, S.D, Norman, B.H. | | Deposit date: | 2020-01-08 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure-based, multi-targeted drug discovery approach to eicosanoid inhibition: Dual inhibitors of mPGES-1 and 5-lipoxygenase activating protein (FLAP).
Biochim Biophys Acta Gen Subj, 1865, 2020
|
|
1O45
 
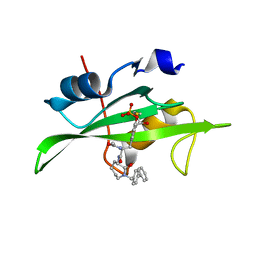 | | CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU84687. | | Descriptor: | N-ACETYL-N-[1-(1,1'-BIPHENYL-4-YLMETHYL)-2-OXOAZEPAN-3-YL]-3-FORMYL-O-PHOSPHONOTYROSINAMIDE, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE SRC | | Authors: | Lange, G, Loenze, P, Liesum, A. | | Deposit date: | 2003-06-15 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Requirements for specific binding of low affinity inhibitor fragments to the SH2 domain of (pp60)Src are identical to those for high affinity binding of full length inhibitors.
J.Med.Chem., 46, 2003
|
|
4ML4
 
 | | Crystal structure of the complex of signaling glycoprotein from buffalo (SPB-40) with tetrahydropyran at 2.5 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3-like protein 1, TETRAHYDROPYRAN | | Authors: | Yamini, S, Chaudhary, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-09-06 | | Release date: | 2013-09-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the complex of signaling glycoprotein from buffalo (SPB-40) with tetrahydropyran at 2.5 A resolution
To be Published
|
|
1O4F
 
 | | CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU79073. | | Descriptor: | 1,2,3,4-TETRAHYDROQUINOLIN-8-YL DIHYDROGEN PHOSPHATE, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE SRC | | Authors: | Lange, G, Loenze, P, Liesum, A. | | Deposit date: | 2003-06-15 | | Release date: | 2004-02-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Requirements for specific binding of low affinity inhibitor fragments to the SH2 domain of (pp60)Src are identical to those for high affinity binding of full length inhibitors.
J.Med.Chem., 46, 2003
|
|
6EUO
 
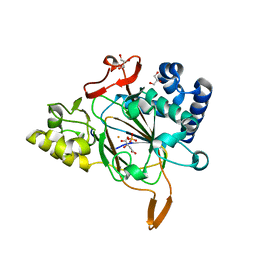 | | Crystal structure of APO Fe(II)/alpha-ketoglutarate dependent dioxygenase KDO5 | | Descriptor: | D-MALATE, FE (III) ION, GLYCEROL, ... | | Authors: | Isabet, T, Stura, E, Legrand, P, Zaparucha, A, Bastard, K. | | Deposit date: | 2017-10-30 | | Release date: | 2018-11-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Studies based on two Lysine Dioxygenases with Distinct Regioselectivity Brings Insights Into Enzyme Specificity within the Clavaminate Synthase-Like Family.
Sci Rep, 8, 2018
|
|
7AI2
 
 | |
3J9I
 
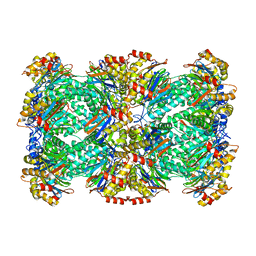 | | Thermoplasma acidophilum 20S proteasome | | Descriptor: | Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Li, X, Mooney, P, Zheng, S, Booth, C, Braunfeld, M.B, Gubbens, S, Agard, D.A, Cheng, Y. | | Deposit date: | 2015-02-02 | | Release date: | 2015-02-18 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM.
Nat.Methods, 10, 2013
|
|
1O4K
 
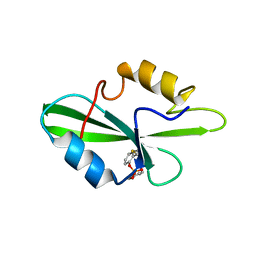 | | CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH PASBN. | | Descriptor: | BENZYL 2-FORMYLPHENYL HYDROGEN PHOSPHATE, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE SRC | | Authors: | Lange, G, Loenze, P, Liesum, A. | | Deposit date: | 2003-06-15 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Requirements for specific binding of low affinity inhibitor fragments to the SH2 domain of (pp60)Src are identical to those for high affinity binding of full length inhibitors.
J.Med.Chem., 46, 2003
|
|
6BG0
 
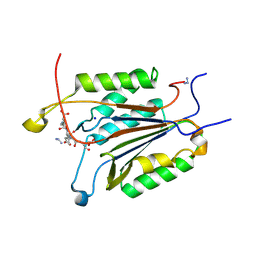 | | Caspase-3 Mutant - D9A,D28A,S150D | | Descriptor: | AC-ASP-GLU-VAL-ASP-CMK, AZIDE ION, Caspase-3, ... | | Authors: | Thomas, M.E, Grinshpon, R, Swartz, P.D, Clark, A.C. | | Deposit date: | 2017-10-27 | | Release date: | 2018-02-21 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (2.125 Å) | | Cite: | Modifications to a common phosphorylation network provide individualized control in caspases.
J. Biol. Chem., 293, 2018
|
|
6EV6
 
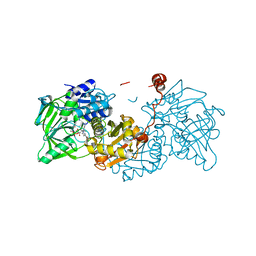 | | Structure of E282Q A. niger Fdc1 with prFMN in the hydroxylated and ketimine forms | | Descriptor: | 1-deoxy-5-O-phosphono-1-[(10aR)-2,2,3,4-tetramethyl-8,10-dioxo-1,2,8,9,10,10a-hexahydro-6H-indeno[1,7-ef]pyrimido[4,5-b][1,4]diazepin-6-yl]-D-ribitol, Ferulic acid decarboxylase 1, MANGANESE (II) ION, ... | | Authors: | Bailey, S.S, David, L, Payne, K.A.P. | | Deposit date: | 2017-11-01 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The role of conserved residues in Fdc decarboxylase in prenylated flavin mononucleotide oxidative maturation, cofactor isomerization, and catalysis.
J. Biol. Chem., 293, 2018
|
|
6EXH
 
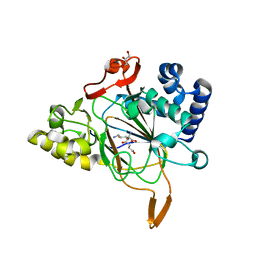 | | Crystal structure of the complex Fe(II)/alpha-ketoglutarate dependent dioxygenase KDO5 with Fe(II)/succinate/(4R)-4-hydroxy-L-lysine | | Descriptor: | 4-HYDROXY-LYSINE, FE (III) ION, GLYCEROL, ... | | Authors: | Isabet, T, Stura, E, Legrand, P, Zaparucha, A, Bastard, K. | | Deposit date: | 2017-11-08 | | Release date: | 2018-11-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Studies based on two Lysine Dioxygenases with Distinct Regioselectivity Brings Insights Into Enzyme Specificity within the Clavaminate Synthase-Like Family.
Sci Rep, 8, 2018
|
|
7AP7
 
 | | Structure of the W64R amyloidogenic variant of human lysozyme | | Descriptor: | Lysozyme C, SULFATE ION | | Authors: | Vettore, N, Herman, R, Kerff, F, Charlier, P, Sauvage, E, Brans, A, Morray, J, Dobson, C, Kumita, J, Dumoulin, M. | | Deposit date: | 2020-10-16 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Characterisation of the structural, dynamic and aggregation properties of the W64R amyloidogenic variant of human lysozyme.
Biophys.Chem., 271, 2021
|
|
4CPG
 
 | |
7STR
 
 | | Crystal Structure of Human Fab S24-1063 in the Complex with the N-teminal Domain of Nucleocapsid Protein from SARS CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, Fab S24-1063, Heavy chain, ... | | Authors: | Kim, Y, Maltseva, N, Tesar, C, Jedrzejczak, R, Dugan, H, Stamper, C, Wilson, P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-11-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Epitopes recognition of SARS-CoV-2 nucleocapsid RNA binding domain by human monoclonal antibodies.
Iscience, 27, 2024
|
|
6PRE
 
 | | SBP RafE in complex with verbascose | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter sugar-binding protein, alpha-D-galactopyranose-(1-6)-alpha-D-galactopyranose-(1-6)-alpha-D-galactopyranose-(1-6)-[beta-D-fructofuranose-(2-1)]alpha-D-glucopyranose | | Authors: | Meier, E.P.W, Boraston, A.B. | | Deposit date: | 2019-07-10 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular analysis of an enigmaticStreptococcus pneumoniaevirulence factor: The raffinose-family oligosaccharide utilization system.
J.Biol.Chem., 294, 2019
|
|
2YOE
 
 | | X-ray structure of a pentameric ligand gated ion channel from Erwinia chrysanthemi (ELIC) in complex with GABA and flurazepam | | Descriptor: | CYS-LOOP LIGAND-GATED ION CHANNEL, Flurazepam, GAMMA-AMINO-BUTANOIC ACID | | Authors: | Spurny, R, Brams, M, Nury, H, Legrand, P, Ulens, C. | | Deposit date: | 2012-10-23 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Pentameric Ligand-Gated Ion Channel Elic is Activated by Gaba and Modulated by Benzodiazepines.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6SZT
 
 | | Crystal structure of YTHDC1 with fragment 10 (DHU_DC1_076) | | Descriptor: | 6-[[methyl(thiophen-3-ylmethyl)amino]methyl]-5~{H}-pyrimidine-2,4-dione, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-02 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
