8SOQ
 
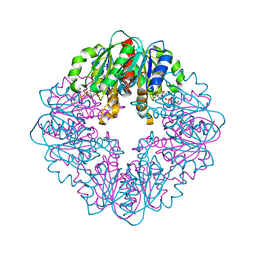 | | S127A variant of LarB, a carboxylase/hydrolase involved in synthesis of the cofactor for lactate racemase, in complex with authentic substrate NaAD | | Descriptor: | MAGNESIUM ION, NICOTINIC ACID ADENINE DINUCLEOTIDE, Pyridinium-3,5-biscarboxylic acid mononucleotide synthase | | Authors: | Chatterjee, S, Rankin, J.A, Hu, J, Hausinger, R.P. | | Deposit date: | 2023-04-29 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the LarB-Substrate Complex and Identification of a Reaction Intermediate during Nickel-Pincer Nucleotide Cofactor Biosynthesis.
Biochemistry, 62, 2023
|
|
5A32
 
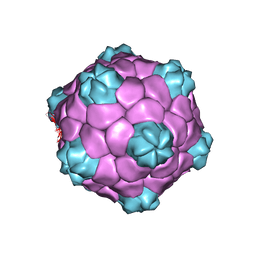 | | Electron cryo-microscopy of Cowpea Mosaic Virus containing RNA-1 (CPMVb) | | Descriptor: | RNA2 POLYPROTEIN | | Authors: | Hesketh, E.L, Meshcheriakova, Y, Dent, K.C, Saxena, P, Thompson, R, Cockburn, J.J, Lomonossoff, G.P, Ranson, N.A. | | Deposit date: | 2015-05-27 | | Release date: | 2015-12-23 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Mechanisms of assembly and genome packaging in an RNA virus revealed by high-resolution cryo-EM.
Nat Commun, 6, 2015
|
|
7B1W
 
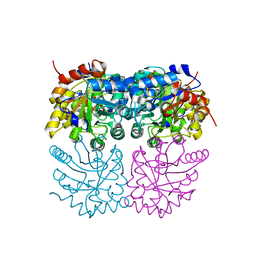 | | Crystal structure of plastidial ribulose epimerase RPE1 from the model alga Chlamydomonas reinhardtii | | Descriptor: | Ribulose-phosphate 3-epimerase, ZINC ION | | Authors: | Henri, J, Zaffagnini, M, Tedesco, D, Crozet, P, Lemaire, S.D. | | Deposit date: | 2020-11-25 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.935 Å) | | Cite: | Characterization of chloroplast ribulose-5-phosphate-3-epimerase from the microalga Chlamydomonas reinhardtii.
Plant Physiol., 2023
|
|
2J6T
 
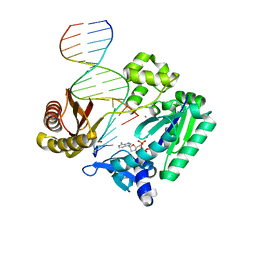 | | Ternary complex of Sulfolobus solfataricus Dpo4 DNA polymerase, O6- methylguanine modified DNA, and dATP. | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP *TP*TP*C)-3', 5'-D(*TP*CP*AP*TP*XP*GP*AP*AP*TP*CP *CP*TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Irimia, A, Eoff, R.L, Guengerich, F.P, Egli, M. | | Deposit date: | 2006-10-04 | | Release date: | 2006-11-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Sulfolobus Solfataricus DNA Polymerase Dpo4 is Partially Inhibited by "Wobble" Pairing between O6- Methylguanine and Cytosine, But Accurate Bypass is Preferred.
J.Biol.Chem., 282, 2007
|
|
6UW7
 
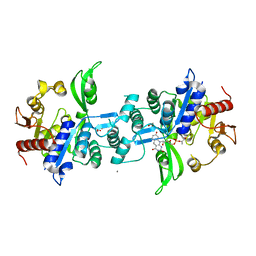 | | The crystal structure of FbiA from Mycobacterium smegmatis, Dehydro-F420-0 bound form | | Descriptor: | 2-[oxidanyl-[(2~{R},3~{S},4~{S})-2,3,4-tris(oxidanyl)-5-[2,4,8-tris(oxidanylidene)-1,9-dihydropyrimido[4,5-b]quinolin-10-yl]pentoxy]phosphoryl]oxyprop-2-enoic acid, CALCIUM ION, GLYCEROL, ... | | Authors: | Grinter, R, Gillett, D, Cordero, P.R.F, Izore, T, Greening, C. | | Deposit date: | 2019-11-04 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.342 Å) | | Cite: | Cellular and Structural Basis of Synthesis of the Unique Intermediate Dehydro-F420-0 in Mycobacteria.
mSystems, 5, 2020
|
|
7XVM
 
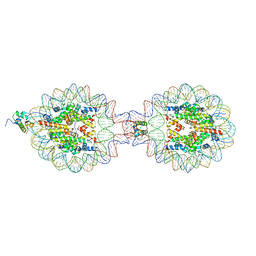 | | Crystal Structure of Nucleosome-H5 Linker Histone Assembly (sticky-169a DNA fragment) | | Descriptor: | CALCIUM ION, CHLORIDE ION, DNA (169-MER), ... | | Authors: | Adhireksan, Z, Qiuye, B, Lee, P.L, Sharma, D, Padavattan, S, Davey, C.A. | | Deposit date: | 2022-05-24 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Crystal Structure of Nucleosome-H1.0 Linker Histone Assembly (sticky-169a DNA fragment)
To Be Published
|
|
5A8F
 
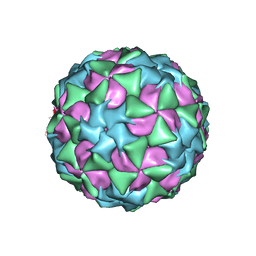 | | Structure and genome release mechanism of human cardiovirus Saffold virus-3 | | Descriptor: | GENOME POLYPHUMAN SAFFOLD VIRUS-3 VP3 PROTEIN, HUMAN SAFFOLD VIRUS-3 VP1, HUMAN SAFFOLD VIRUS-3 VP2 | | Authors: | Mullapudi, E, Novacek, J, Palkova, L, Kulich, P, Lindberg, M, vanKuppeveld, F.J.M, Plevka, P. | | Deposit date: | 2015-07-15 | | Release date: | 2016-06-08 | | Last modified: | 2019-10-30 | | Method: | ELECTRON MICROSCOPY (10.6 Å) | | Cite: | Structure and Genome Release Mechanism of Human Cardiovirus Saffold Virus-3.
J.Virol., 90, 2016
|
|
8TPW
 
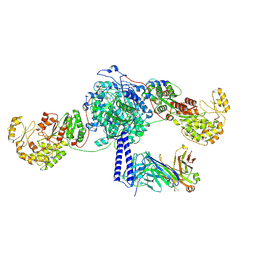 | | Crosslinked 6-deoxyerythronolide B synthase (DEBS) Module 3 in complex with antibody fragment 1B2: cis-oriented 1B2 and ACP | | Descriptor: | Antibody Fragment 1B2, Heavy Chain, Light Chain, ... | | Authors: | Cogan, D.P, Soohoo, A.M, Chen, M, Brodsky, K.L, Liu, Y, Khosla, C. | | Deposit date: | 2023-08-05 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural basis for intermodular communication in assembly-line polyketide biosynthesis.
Nat.Chem.Biol., 2024
|
|
7XVL
 
 | | Crystal Structure of Nucleosome-H1.0 Linker Histone Assembly (sticky-169an DNA fragment) | | Descriptor: | DNA (169-MER), Histone H1.0, Histone H2A type 1-B/E, ... | | Authors: | Adhireksan, Z, Qiuye, B, Lee, P.L, Sharma, D, Padavattan, S, Davey, C.A. | | Deposit date: | 2022-05-24 | | Release date: | 2023-05-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.506 Å) | | Cite: | Crystal Structure of Nucleosome-H1.0 Linker Histone Assembly (sticky-169an DNA fragment)
To Be Published
|
|
2VEU
 
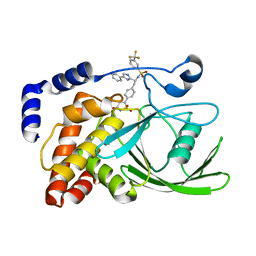 | | Crystal structure of protein tyrosine phosphatase 1B in complex with an isothiazolidinone-containing inhibitor | | Descriptor: | N-[(1S)-2-{4-[(5S)-1,1-dioxido-3-oxoisothiazolidin-5-yl]phenyl}-1-(4-phenyl-1H-imidazol-2-yl)ethyl]-3-(trifluoromethyl)benzenesulfonamide, TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 1 | | Authors: | Douty, B, Wayland, B, Ala, P.J, Bower, M.J, Pruitt, J, Bostrom, L, Wei, M, Klabe, R, Gonneville, L, Wynn, R, Burn, T.C, Liu, P.C.C, Combs, A.P, Yue, E.W. | | Deposit date: | 2007-10-27 | | Release date: | 2007-11-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Isothiazolidinone Inhibitors of Ptp1B Containing Imidazoles and Imidazolines
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2J6S
 
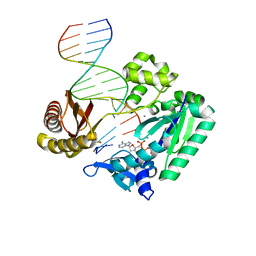 | | Ternary complex of Sulfolobus solfataricus Dpo4 DNA polymerase, O6- methylguanine modified DNA, and dATP. | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP *TP*TP*CP*C)-3', 5'-D(*TP*CP*AP*TP*XP*GP*AP*AP*TP*CP *CP*TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Irimia, A, Eoff, R.L, Guengerich, F.P, Egli, M. | | Deposit date: | 2006-10-04 | | Release date: | 2006-11-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Sulfolobus Solfataricus DNA Polymerase Dpo4 is Partially Inhibited by "Wobble" Pairing between O6- Methylguanine and Cytosine, But Accurate Bypass is Preferred.
J.Biol.Chem., 282, 2007
|
|
6VAF
 
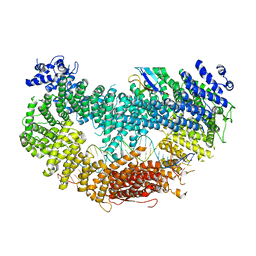 | |
7XX5
 
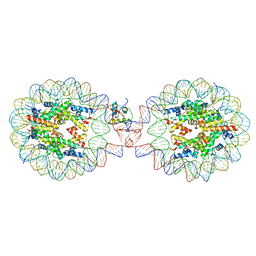 | | Crystal Structure of Nucleosome-H1.3 Linker Histone Assembly (sticky-169a DNA fragment) | | Descriptor: | CALCIUM ION, DNA (169-MER), Histone H1.3, ... | | Authors: | Adhireksan, Z, Qiuye, B, Lee, P.L, Sharma, D, Padavattan, S, Davey, C.A. | | Deposit date: | 2022-05-28 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Crystal Structure of Nucleosome-H1.0 Linker Histone Assembly (sticky-169a DNA fragment)
To Be Published
|
|
5WYQ
 
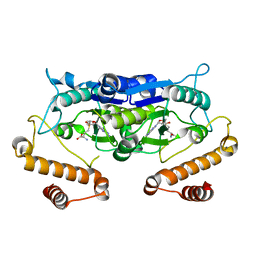 | | Crystal Structure and catalytic mechanism of the essential m1G37 tRNA methyltransferase TrmD from Pseudomonas aeruginosa | | Descriptor: | S-ADENOSYLMETHIONINE, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Jaroensuk, J, Liew, C.W, Atichartpongkul, S, Chionh, Y.H, Wong, Y.H, Zhong, W, McBee, M.E, Maenpuen, S, Prestwich, E.G, DeMott, M.S, Mongkolsuk, S, Dedon, P.C, Lescar, J, Fuangthong, M, Chaiyen, P. | | Deposit date: | 2017-01-15 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure and catalytic mechanism of the essential m1G37 tRNA methyltransferase TrmD fromPseudomonas aeruginosa.
Rna, 2019
|
|
1KD1
 
 | | Co-crystal Structure of Spiramycin bound to the 50S Ribosomal Subunit of Haloarcula marismortui | | Descriptor: | 23S RRNA, 5S RRNA, CADMIUM ION, ... | | Authors: | Hansen, J.L, Ippolito, J.A, Ban, N, Nissen, P, Moore, P.B, Steitz, T.A. | | Deposit date: | 2001-11-12 | | Release date: | 2002-07-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structures of four macrolide antibiotics bound to the large ribosomal subunit.
Mol.Cell, 10, 2002
|
|
1UJL
 
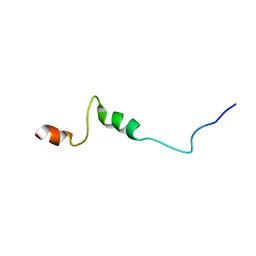 | | Solution Structure of the HERG K+ channel S5-P extracellular linker | | Descriptor: | Potassium voltage-gated channel subfamily H member 2 | | Authors: | Torres, A.M, Bansal, P.S, Sunde, M, Clarke, C.E, Bursill, J.A, Smith, D.J, Bauskin, A, Breit, S.N, Campbell, T.J, Alewood, P.F, Kuchel, P.W, Vandenberg, J.I. | | Deposit date: | 2003-08-05 | | Release date: | 2003-11-04 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of the HERG K+ channel S5P extracellular linker: role of an amphipathic alpha-helix
in C-type inactivation.
J.Biol.Chem., 278, 2003
|
|
8JD9
 
 | | Cyro-EM structure of the Na+/H+ antipoter SOS1 from Arabidopsis thaliana,class1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Sodium/hydrogen exchanger 7 | | Authors: | Yang, G.H, Zhang, Y.M, Zhou, J.Q, Jia, Y.T, Xu, X, Fu, P, Wu, H.Y. | | Deposit date: | 2023-05-13 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Structural basis for the activity regulation of Salt Overly Sensitive 1 in Arabidopsis salt tolerance.
Nat.Plants, 9, 2023
|
|
4A3D
 
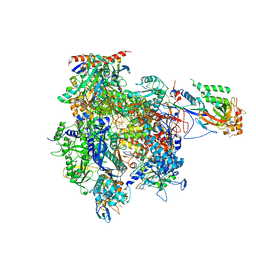 | | RNA Polymerase II initial transcribing complex with a 6nt DNA-RNA hybrid | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB1, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB11, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB2, ... | | Authors: | Cheung, A.C.M, Sainsbury, S, Cramer, P. | | Deposit date: | 2011-09-30 | | Release date: | 2011-12-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis of initial RNA polymerase II transcription.
EMBO J., 30, 2011
|
|
4A3C
 
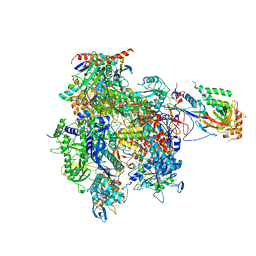 | | RNA Polymerase II initial transcribing complex with a 5nt DNA-RNA hybrid | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB1, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB11, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB2, ... | | Authors: | Cheung, A.C.M, Sainsbury, S, Cramer, P. | | Deposit date: | 2011-09-30 | | Release date: | 2011-11-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis of initial RNA polymerase II transcription.
EMBO J., 30, 2011
|
|
5OO2
 
 | | Crystal structure of Mycolicibacterium hassiacum glucosylglycerate hydrolase (MhGgH) E419A variant in complex with glucosylglycolate | | Descriptor: | (alpha-D-glucopyranosyloxy)acetic acid, GLYCEROL, SERINE, ... | | Authors: | Cereija, T.B, Macedo-Ribeiro, S, Pereira, P.J.B. | | Deposit date: | 2017-08-05 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | The structural characterization of a glucosylglycerate hydrolase provides insights into the molecular mechanism of mycobacterial recovery from nitrogen starvation.
Iucrj, 6, 2019
|
|
2P6B
 
 | | Crystal Structure of Human Calcineurin in Complex with PVIVIT Peptide | | Descriptor: | CALCIUM ION, Calcineurin subunit B isoform 1, Calmodulin-dependent calcineurin A subunit alpha isoform, ... | | Authors: | Li, H, Zhang, L, Rao, A, Harrison, S.C, Hogan, P.G. | | Deposit date: | 2007-03-16 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of calcineurin in complex with PVIVIT peptide: Portrait of a low-affinity signalling interaction
J.Mol.Biol., 369, 2007
|
|
8J5J
 
 | | The crystal structure of bat coronavirus RsYN04 RBD bound to the antibody S43 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, nano antibody S43 | | Authors: | Zhao, R.C, Niu, S, Han, P, Qi, J.X, Gao, G.F, Wang, Q.H. | | Deposit date: | 2023-04-23 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cross-species recognition of bat coronavirus RsYN04 and cross-reaction of SARS-CoV-2 antibodies against the virus.
Zool.Res., 44, 2023
|
|
5WO3
 
 | | Chaperone Spy bound to Im7 (Im7 un-modeled) | | Descriptor: | CHLORIDE ION, IMIDAZOLE, Periplasmic chaperone Spy, ... | | Authors: | Horowitz, S, Koldewey, P, Martin, R, Bardwell, J.C.A. | | Deposit date: | 2017-08-01 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Visualizing chaperone-assisted protein folding.
Nat. Struct. Mol. Biol., 23, 2016
|
|
7K8T
 
 | | Structure of the SARS-CoV-2 S 6P trimer in complex with the human neutralizing antibody Fab fragment, C002 (State 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C002 Fab Heavy Chain, ... | | Authors: | Malyutin, A.G, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
6M7E
 
 | | Structure of bovine lactoperoxidase with multiple iodide ions in the distaline heme cavity. | | Descriptor: | 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Maurya, A, Viswanathan, V, Pandey, N, Singh, A.K, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2020-03-18 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structure of bovine lactoperoxidase with multiple iodide ions in the distaline heme cavity.
To Be Published
|
|
