6P0R
 
 | | Methyltransferase domain of human suppressor of variegation 3-9 homolog 2 (SUV39H2) in complex with OTS186935 inhibitor | | Descriptor: | (3S)-1-[2-(5-chloro-2,4-dimethoxyphenyl)imidazo[1,2-a]pyridin-7-yl]-N-[(pyridin-4-yl)methyl]pyrrolidin-3-amine, Histone-lysine N-methyltransferase SUV39H2, UNKNOWN ATOM OR ION, ... | | Authors: | Halabelian, L, Dong, A, Zeng, H, Loppnau, P, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-05-17 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Methyltransferase domain of human suppressor of variegation 3-9 homolog 2 (SUV39H2) in complex with OTS186935 inhibitor
to be published
|
|
6PL5
 
 | | Structural coordination of polymerization and crosslinking by a peptidoglycan synthase complex | | Descriptor: | Penicillin-binding protein 2/cell division protein FtsI, Peptidoglycan glycosyltransferase RodA, Unknown peptide | | Authors: | Sjodt, M, Rohs, P.D.A, Erlandson, S.C, Zheng, S, Rudner, D.Z, Bernhardt, T.G, Kruse, A.C. | | Deposit date: | 2019-06-30 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural coordination of polymerization and crosslinking by a SEDS-bPBP peptidoglycan synthase complex.
Nat Microbiol, 5, 2020
|
|
5LV1
 
 | | 2.12 A resolution structure of PtxB from Prochlorococcus marinus (MIT 9301) in complex with phosphite | | Descriptor: | PtxB, oxidanylphosphinate | | Authors: | Bisson, C, Adams, N.B.P, Polyviou, D, Bibby, T.S, Hunter, C.N, Hitchcock, A. | | Deposit date: | 2016-09-12 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | The molecular basis of phosphite and hypophosphite recognition by ABC-transporters.
Nat Commun, 8, 2017
|
|
7SNS
 
 | | 1.55A Resolution Structure of NanoLuc Luciferase | | Descriptor: | ACETATE ION, Oplophorus-luciferin 2-monooxygenase catalytic subunit | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Wood, M.G, Encell, L.P, Wood, K.V. | | Deposit date: | 2021-10-28 | | Release date: | 2022-11-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | 1.55A Resolution Structure of NanoLuc Luciferase
To be published
|
|
5LYS
 
 | | The crystal structure of 7SK 5'-hairpin - Gold derivative | | Descriptor: | 7SK RNA, GOLD ION, MAGNESIUM ION, ... | | Authors: | Martinez-Zapien, D, Legrand, P, McEwen, A.G, Pasquali, S, Dock-Bregeon, A.-C. | | Deposit date: | 2016-09-28 | | Release date: | 2017-01-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | The crystal structure of the 5 functional domain of the transcription riboregulator 7SK.
Nucleic Acids Res., 45, 2017
|
|
6SHX
 
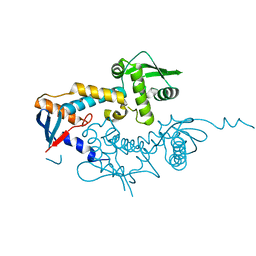 | | DNA mismatch repair proteins MLH1 and MLH3 | | Descriptor: | DNA mismatch repair protein MLH1, DNA mismatch repair protein MLH3, ZINC ION | | Authors: | Dai, J, Chervy, P, Legrand, P, Ropars, V, Charbonnier, J.B. | | Deposit date: | 2019-08-08 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis of the dual role of the Mlh1-Mlh3 endonuclease in MMR and in meiotic crossover formation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6SNS
 
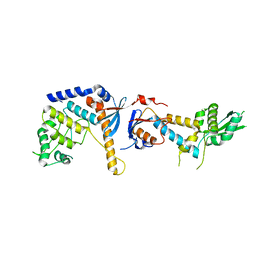 | | DNA mismatch repair proteins MLH1 and MLH3 | | Descriptor: | DNA mismatch repair protein MLH1, DNA mismatch repair protein MLH3, ZINC ION | | Authors: | Dai, J, Chervy, P, Legrand, P, Ropars, V, Charbonnier, J.B. | | Deposit date: | 2019-08-27 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular basis of the dual role of the Mlh1-Mlh3 endonuclease in MMR and in meiotic crossover formation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6SNV
 
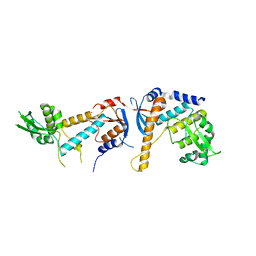 | | DNA mismatch repair proteins MLH1 and MLH3 | | Descriptor: | DNA mismatch repair protein MLH1, DNA mismatch repair protein MLH3, ZINC ION | | Authors: | Dai, J, Chervy, P, Legrand, P, Ropars, V, Charbonnier, J.B. | | Deposit date: | 2019-08-27 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis of the dual role of the Mlh1-Mlh3 endonuclease in MMR and in meiotic crossover formation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7N3E
 
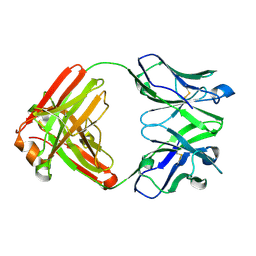 | | Crystal structure of an anti-SARS-CoV-2 human neutralizing antibody Fab fragment C032 | | Descriptor: | C032 Fab Heavy Chain, C032 Fab Light Chain | | Authors: | Flyak, A.I, Bjorkman, P.J, Barnes, C.O. | | Deposit date: | 2021-06-01 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations.
Immunity, 54, 2021
|
|
6VMG
 
 | | Chloroplast ATP synthase (O3, CF1FO) | | Descriptor: | ATP synthase delta chain, chloroplastic, ATP synthase epsilon chain, ... | | Authors: | Yang, J.-H, Williams, D, Kandiah, E, Fromme, P, Chiu, P.-L. | | Deposit date: | 2020-01-27 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.46 Å) | | Cite: | Structural basis of redox modulation on chloroplast ATP synthase.
Commun Biol, 3, 2020
|
|
6VM5
 
 | | Structure of Moraxella osloensis Cap4 SAVED/CARF-domain containing receptor | | Descriptor: | MAGNESIUM ION, SAVED domain-containing protein | | Authors: | Lowey, B, Whiteley, A.T, Keszei, A.F.A, Morehouse, B.R, Antine, S.P, Cabrera, V, Schwede, F, Mekalanos, J.J, Shao, S, Lee, A.S.Y, Kranzusch, P.J. | | Deposit date: | 2020-01-27 | | Release date: | 2020-06-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | CBASS Immunity Uses CARF-Related Effectors to Sense 3'-5'- and 2'-5'-Linked Cyclic Oligonucleotide Signals and Protect Bacteria from Phage Infection.
Cell, 182, 2020
|
|
6VOK
 
 | | Chloroplast ATP synthase (R3, CF1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase delta chain, ... | | Authors: | Yang, J.-H, Williams, D, Kandiah, E, Fromme, P, Chiu, P.-L. | | Deposit date: | 2020-01-30 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Structural basis of redox modulation on chloroplast ATP synthase.
Commun Biol, 3, 2020
|
|
7NVV
 
 | | XPB-containing part of TFIIH in a post-translocated state (with ADP-BeF3) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, General transcription and DNA repair factor IIH helicase subunit XPB, ... | | Authors: | Aibara, S, Schilbach, S, Cramer, P. | | Deposit date: | 2021-03-16 | | Release date: | 2021-05-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of mammalian RNA polymerase II pre-initiation complexes.
Nature, 594, 2021
|
|
7NSY
 
 | | Drosophila PGRP-LB C160S mutant | | Descriptor: | Isoform A of Peptidoglycan-recognition protein LB | | Authors: | Orlans, J, Aller, P, Da Silva, P. | | Deposit date: | 2021-03-08 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | PGRP-LB: An Inside View into the Mechanism of the Amidase Reaction.
Int J Mol Sci, 22, 2021
|
|
5HP2
 
 | | Human Adenosine Deaminase Acting on dsRNA (ADAR2) bound to dsRNA sequence derived from S. cerevisiae BDF2 gene with AU basepair at reaction site | | Descriptor: | Double-stranded RNA-specific editase 1, INOSITOL HEXAKISPHOSPHATE, RNA (5'-R(*GP*AP*CP*UP*GP*AP*AP*CP*GP*AP*CP*UP*AP*AP*UP*GP*UP*GP*GP*GP*GP*AP*A)-3'), ... | | Authors: | Matthews, M.M, Fisher, A.J, Beal, P.A. | | Deposit date: | 2016-01-20 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.983 Å) | | Cite: | Structures of human ADAR2 bound to dsRNA reveal base-flipping mechanism and basis for site selectivity.
Nat.Struct.Mol.Biol., 23, 2016
|
|
7NSZ
 
 | | Drosophila PGRP-LB Y78F mutant | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Isoform A of Peptidoglycan-recognition protein LB, SODIUM ION, ... | | Authors: | Orlans, J, Aller, P, Da Silva, P. | | Deposit date: | 2021-03-08 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | PGRP-LB: An Inside View into the Mechanism of the Amidase Reaction.
Int J Mol Sci, 22, 2021
|
|
1B39
 
 | | HUMAN CYCLIN-DEPENDENT KINASE 2 PHOSPHORYLATED ON THR 160 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PROTEIN (CELL DIVISION PROTEIN KINASE 2) | | Authors: | Brown, N.R, Noble, M.E.M, Lawrie, A.M, Morris, M.C, Tunnah, P, Divita, G, Johnson, L.N, Endicott, J.A. | | Deposit date: | 1998-12-17 | | Release date: | 1998-12-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Effects of phosphorylation of threonine 160 on cyclin-dependent kinase 2 structure and activity.
J.Biol.Chem., 274, 1999
|
|
5MMO
 
 | | E. coli DNA Gyrase B 24 kDa ATPase domain in complex with [3-(3-ethyl-ureido)-5-(pyridin-4-yl)-isoquinolin-8-yl-methyl]-carbamic acid prop-2-ynyl ester | | Descriptor: | DNA gyrase subunit B, PHOSPHATE ION, prop-2-ynyl ~{N}-[[3-(ethylcarbamoylamino)-5-pyridin-4-yl-isoquinolin-8-yl]methyl]carbamate | | Authors: | Panchaud, P, Bruyere, T, Blumstein, A.-C, Bur, D, Chambovey, A, Ertel, E.A, Gude, M, Hubschwerlen, C, Jacob, L, Kimmerlin, T, Pfeifer, T, Prade, L, Seiler, P, Ritz, D, Rueedi, G. | | Deposit date: | 2016-12-12 | | Release date: | 2017-04-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Discovery and Optimization of Isoquinoline Ethyl Ureas as Antibacterial Agents.
J. Med. Chem., 60, 2017
|
|
5MY1
 
 | | E. coli expressome | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Kohler, R, Mooney, R.A, Mills, D.J, Kostrewa, D, Landick, R, Cramer, P. | | Deposit date: | 2017-01-25 | | Release date: | 2017-04-26 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Architecture of a transcribing-translating expressome.
Science, 356, 2017
|
|
7NT0
 
 | | Drosophila PGRP-LB Y78F mutant in complex with tracheal cytotoxin (TCT) | | Descriptor: | GLCNAC(BETA1-4)-MURNAC(1,6-ANHYDRO)-L-ALA-GAMMA-D-GLU-MESO-A2PM-D-ALA, Isoform A of Peptidoglycan-recognition protein LB, ZINC ION | | Authors: | Orlans, J, Aller, P, Da Silva, P. | | Deposit date: | 2021-03-08 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | PGRP-LB: An Inside View into the Mechanism of the Amidase Reaction.
Int J Mol Sci, 22, 2021
|
|
6SNT
 
 | | Yeast 80S ribosome stalled on SDD1 mRNA. | | Descriptor: | 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, 40S ribosomal protein S10-A, ... | | Authors: | Tesina, P, Buschauer, R, Cheng, J, Becker, T, Beckmann, R. | | Deposit date: | 2019-08-27 | | Release date: | 2020-03-04 | | Last modified: | 2020-04-22 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | RQT complex dissociates ribosomes collided on endogenous RQC substrate SDD1.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5ILX
 
 | | Crystal structure of Ribosome inactivating protein from Momordica balsamina with Uracil at 1.70 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Ribosome inactivating protein, ... | | Authors: | Singh, P.K, Singh, A, Pandey, S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-03-05 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Ribosome inactivating protein from Momordica balsamina with Uracil at 1.70 Angstrom resolution
To Be Published
|
|
7ZW4
 
 | | Crystal structure of Talin R7R8 domains with Caskin-2 LD-peptide | | Descriptor: | Caskin-2, Talin-1 | | Authors: | Celie, P.H.N, Joosten, R.P, Sonnenberg, A, Perrakis, A. | | Deposit date: | 2022-05-18 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Caskin2 is a novel talin- and Abi1-binding protein that promotes cell motility.
J.Cell.Sci., 137, 2024
|
|
6VVR
 
 | |
7OH5
 
 | | Cryo-EM structure of Drs2p-Cdc50p in the E1-AlFx-ADP state | | Descriptor: | (2R)-1-{[(R)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Timcenko, M, Dieudonne, T, Montigny, C, Boesen, T, Lyons, J.A, Lenoir, G, Nissen, P. | | Deposit date: | 2021-05-09 | | Release date: | 2021-06-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of substrate-independent phosphorylation in a P4-ATPase lipid flippase
J.Mol.Biol., 2021
|
|
